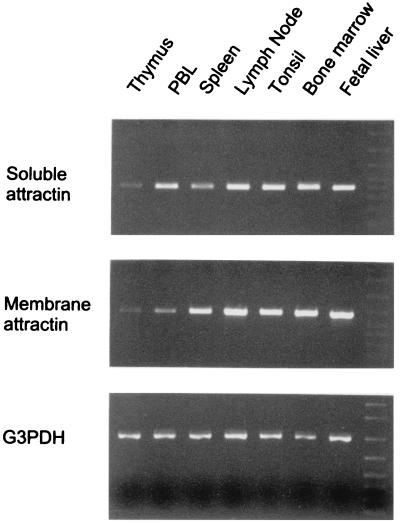
Figure 5.
First-strand cDNA immune tissue [primed from the poly(A) tail] that had been normalized for four housekeeping genes was tested for representation of soluble and membrane attractin. The lower panels depict amplification of a G3PDH (1 kb) fragment for all of the samples tested. Soluble attractin (412 bp) was amplified by using the primers depicted in Fig. 1B. Membrane attractin (668 bp) was amplified by using primers within the 3′ UTR. Identical results were obtained by using the primers designed from coding sequence (552 bp) but required five additional cycles to obtain a strong signal. This is a result of first-strand priming from poly(A) and the length of the membrane attractin 3′ UTR (>4 kb) leading to under-representation of the coding sequence.