Fig. 2.
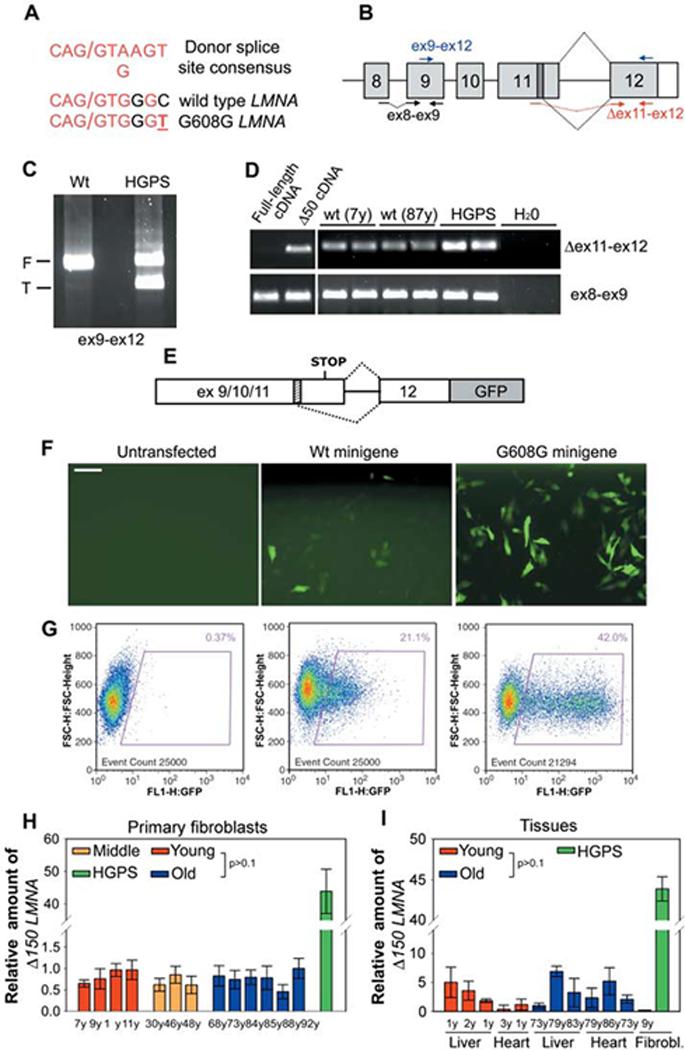
Sporadic use of exon 11 LMNA cryptic splice site in cells from healthy individuals. (A)Comparison between the consensus donor splice sequence, the wild-type LMNA cryptic splice site in exon 11, and the mutated cryptic splice site in HGPS patients. Forward slash indicates the splice junction. (B)Schematic representation of splicing events (dotted black lines) involving LMNA exon 11 and exon 12. The cryptic splice site (dark box) and the position of different primer sets used for RT-PCR are indicated. RT-PCR analysis of wild-type and HGPS fibroblasts using (C) primers detecting both the full-length (F) and the truncated (T) LMNA isoforms or (D) primers specific for Δ150 LMNA (Δ ex11-ex12). Control ex8-ex9 primers detect all LMNA transcripts. Samples were analyzed in duplicate. Plasmids containing either full-length or truncated lamin A cDNA were used as controls for specificity of Δ150 LMNA detection. (E) Schematic representation of LMNA splicing reporter (9). The cryptic splice site (dashed box) is indicated. (F) Immunofluorescence microscopy and (G) fluorescence-activated cell sorting analysis of fibroblasts from a healthy individual transiently transfected with the indicated reporter minigene or untransfected. Scale bar, 60 μm. Quantitative RT-PCR analysis of (H) wild-type fibroblasts and (I) tissues from healthy individuals of indicated age and a HGPS patient. Δ150 LMNA RNA detected by using Δex11-ex12 primers relative to total LMNA RNA detected by using ex8-ex9 primers is shown.
