Figure 2.
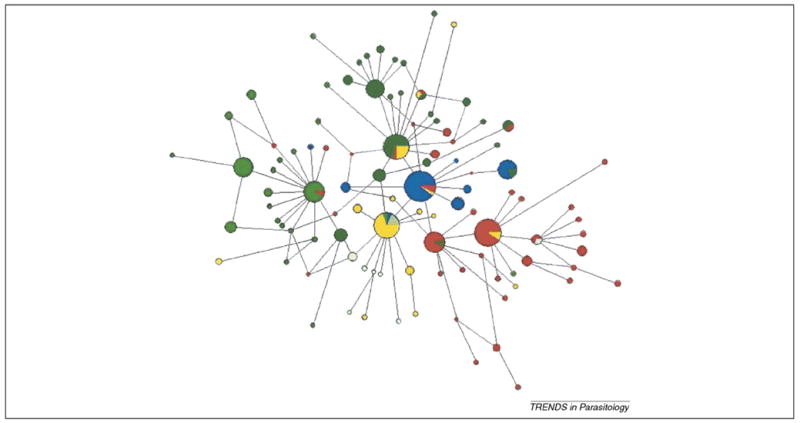
Haplotype network showing the relationships among Plasmodium vivax populations. Haplotype network of the 282 mitochondrial genomes (joint datasets) inferred under a median-joining algorithm with posterior pruning using maximum parsimony criteria as implemented in Network 4.1.1.2 [32]. The size of the circles is proportional to the haplotype frequency, with each color indicating the geographic origin of the sample. The Asian populations (dark green) include isolates from China, Thailand, Indonesia and Vietnam. There are 45 haplotypes in 114 sequences from this region, with a haplotype diversity of 0.948. The Melanesian sample (red) includes 32 haplotypes from 73 sequences, with a haplotype diversity of 0.889; the isolates are from Papua New Guinea, Salomon Islands and Vanuatu. The India–Middle-East sample (yellow) includes 16 haplotypes out of 35 sequenced and a haplotype diversity of 0.881. There is more haplotype diversity in Asia and Melanesia than in any other region considered in this analysis. The population from the Americas (blue) has a small number of haplotypes (seven out of 47 sequences and a haplotype diversity of 0.645), smaller, even, than the African population (light green) (eight haplotypes in 12 sequences, with a haplotype diversity of 0.894).
