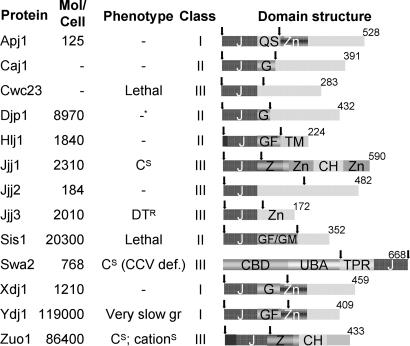
Fig. 1.
Cytosolic J proteins of S. cerevisiae. Molecules per cell are as reported by Ghaemmaghami et al. (11). No data available is indicated by −. Phenotypes tested are as follows: cold sensitivity (CS), resistance to diphtheria toxin (DTR), very compromised for growth (very slow gr.), sensitive to cations (cationS), and defective in uncoating CCVs (CCV def.). Information was obtained from the Saccharomyces Genome database, www.yeastgenome.org. J protein classes were taken from Walsh et al. (7). In the domain structure, arrows indicate the J domain fragments used (Apj11–161, Caj11–139, Cwc231–139, Djp11–127, Hlj11–179, Jjj11–128, Jjj21–132, Jjj31–124, Sis11–167, Swa2362–668, Xdj11–146, Ydj11–134, and Zuo11–234). Numbers indicate amino acids in each full-length J protein. Regions in which these amino acids are highly represented include QS, G, GF, and GF/GM (listed by their standard single-letter amino acid code). J, J domain; Zn, zinc finger; CBD, clathrin-binding domain; UBA, ubiquitin association; TPR, tetratricopeptide repeat; Z, zuotin-like; CH, charged; TM, transmembrane.