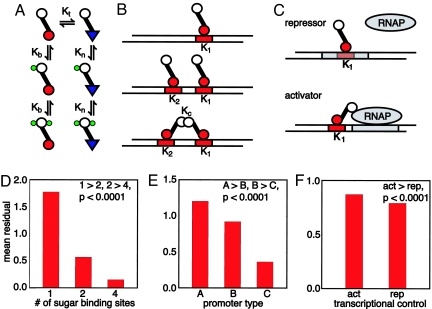
Fig. 2.
A comparison of different regulatory mechanisms for solving the two-state discrimination problem; highly cooperative, negatively controlled genetic networks perform the most accurate inference. (A) The Monod–Changeux–Wyman model of an allosteric transcription factor. Association constants are denoted by Ks. The protein flips between DNA binding (red circles) and non-DNA binding forms (blue triangles). If Kb ≫ Kn, sugar stabilizes the DNA binding state. Conversely, if Kn ≫ Kb, the non-DNA binding state is stabilized. Two sugar binding sites are shown, but we also test models with one and four binding sites. (B) We consider three different promoters: type A, one active operator site (Top); type B, two active operator sites, but with no cooperative binding between transcription factors (Middle); and type C, one active and one inactive operator with cooperative transcription factor binding (Bottom). (C) Transcription can by regulated either negatively, via repressors that obstruct RNA polymerase (RNAP) binding, or positively, via activators that help stabilize RNAP binding. The RNAP binding site (sigma site) is shown in gray, operators in red. (D) Mean residuals (a high residual implies a poor fit) from fits to 50 different posterior probabilities for the models grouped by the numbers of sugars bound by transcription factor. Models with four transcription binding sites perform the best inference (P value for one model type consistently performing better than the other is given; see SI Appendix). (E) Mean residuals for models grouped by promoter type. Cooperative promoters perform best (type C). (F) Mean residuals for models grouped by their mode of transcriptional control. Repressors perform better than activators (for >70% of the fits, corresponding to a P value substantially <10−4).