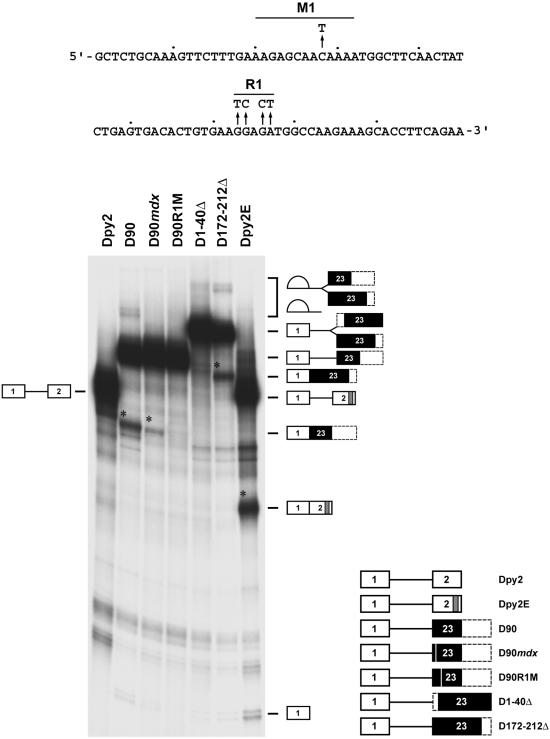
Figure 1. Exon 23 contains purine-rich elements that activate splicing of a heterologous substrate containing a weak 3′ splice site.
The sequence of the first 90 nucleotides of dystrophin exon 23 is shown at the top of the figure. Arrows identify the natural mutation (C→T) occurring in the mdx mouse and the mutations that disrupt the purine-rich region R1. The borders of the M1 element are defined by the homology (12 of 12 positions) shared with the human dystrophin exon 23. Different regions of dystrohin exon 23 were inserted in the second exon of Dpy2, an enhancer-dependent mRNA containing a weak pyrimidine stretch [60]. The structure of the chimeric pre-mRNA substrates subjected to in vitro splicing reactions is shown at the bottom right of the figure. White boxes indicate Dpy2 exons, black boxes indicate different regions of the dystrophin exon 23, dashed boxes indicate portions of exon 23 not included in the substrates, white vertical lines indicate the mdx and R1 mutations. The gray box in Dpy2E identifies the two splicing enhancers found in the α-tropomyosin exon 2 gene [60]. The structures and the mobility of the products and intermediates of splicing are shown on the sides of the gel. D172-212Δ RNA was used as a positive control for D1-40Δ to rule out any exon size inhibitory effect on splicing. Asterisks indicate the position of the spliced RNAs. Splicing reactions, carried out for 70 min at 30°C, were resolved on a 12% denaturing polyacrylamide gel. Comparable results were obtained in three independent experiments.