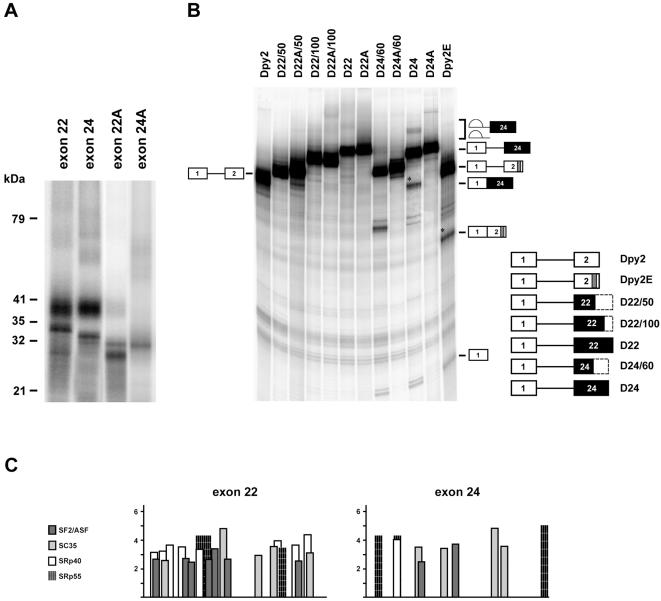
Figure 5. SR proteins interact with dystrophin exon 22 and 24.
A) Different SR proteins associate with exons 22 and 24. 32P labeled exon 22 and 24 RNAs, incubated with SR proteins were subjected to UV-crosslinking as described in Figure 3. Molecular weight markers are shown at the left; exon 22A and 24A correspond to the antisense RNAs used as controls. B) Exon 24, but not exon 22 activates splicing of the enhancer-dependent Dpy2 RNA. Different regions of dystrohin exon 22 (145 nt) and 24 (113 nt) were cloned in the second exon of the enhancer-dependent Dpy2 substrate. The structure of the chimeric constructs subjected to in vitro splicing reactions is shown on the right of the panel, with black boxes indicating exon 22 and 24 and dashed boxes the portion of the corresponding exon not included in the pre-mRNA. A indicates the anti-sense orientation of each region, while numbers after the slash indicate their nucleotide length. Dpy2 and Dpy2E are defined as in Figure 1. The structures and the mobility of the products and intermediates of splicing are shown on both sides of the panel. Asterisks indicate the position of the spliced RNAs. The molecular weigh of the band detected below the D24/60 RNA precursor does not correspond to the expected splicing product. In vitro splicing reactions were carried out as in Figure 1. Comparable results were obtained in two independent experiments. C) Schematic representation of SR binding sites within dystrophin exon 22 and 24 computed by ESEfinder [61], [62].