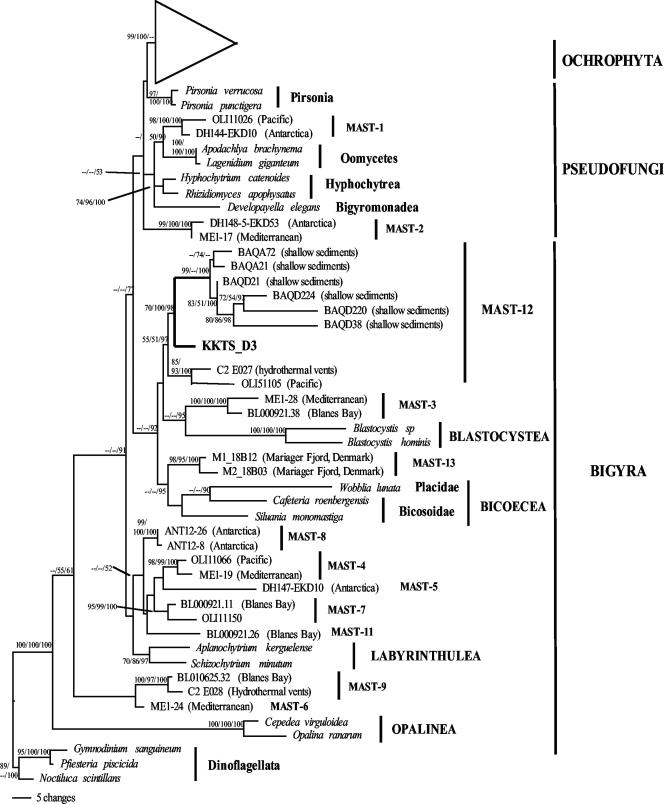
FIG. 1.
Bayesian inference tree for stramenopile 18S rRNA gene sequences, showing the phylogenetic position of clone KKTS_D3 retrieved from an enrichment sample of an oxygen-depleted microbial mat from a small Norwegian estuary. This clone branches within the environment-specific MAST-12 clade. The tree is based on 1,600,000 generations with two runs (each run with three hot chains and one cold chain) using a general time-reversible model. The gamma shape parameter, the rate of invariable sites, the rate matrix, and the base frequencies of the substitution model were estimated by MrBayes (35). The sample frequency was set at 1,000, resulting in 1,601 trees. The numbers at the nodes are ML bootstrap/evolutionary distance bootstrap/Bayesian posterior probability values. For ML and evolutionary distance trees, we used the TrN substitution model with a gamma shape parameter of 0.6482, a rate of invariable sites of 0.3339, and base frequencies and a rate matrix suggested by Modeltest (32). The tree is based on 906 unambiguously aligned positions.