Abstract
Kpp95, isolated on Klebsiella pneumoniae, is a bacteriophage with the morphology of T4-type phages and is capable of rapid lysis of host cells. Its double-stranded genomic DNA (ca. 175 kb, estimated by pulsed-field gel electrophoresis) can be cut only by restriction endonucleases with a cleavage site flanked either by A and T or by T, as tested, suggesting that it contains the modified derivative(s) of G and/or C. Over 26 protein bands were visualized upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the virion proteins. N-terminal sequencing indicated that the most abundant band (46 kDa) is the major coat protein (gp23) which has been cleaved from a signal peptide likely with a length similar to that of T4. Phylogenetic analyses based on the sequences of the central region (263 amino acid residues) of gp23 and the full length of gp18 and gp19 placed Kpp95 among the pseudo-T-even subgroup, most closely related to the coliphage JS98. In addition to being able to lyse many extended-spectrum β-lactamase strains of K. pneumoniae, Kpp95 can lyse Klebsiella oxytoca, Enterobacter agglomerans, and Serratia marcescens cells. Thus, Kpp95 deserves further studies for development as a component of a therapeutic cocktail, owing to its high efficiencies of host lysis plus extended host range.
The Klebsiella spp., ubiquitous in nature, are opportunistic human pathogens attacking primarily immunocompromised individuals who are hospitalized and suffering from severe underlying diseases such as diabetes mellitus or chronic pulmonary obstruction. It is estimated that Klebsiella spp. cause 8% of nosocomial bacterial infections in the United States and in Europe, placing these bacteria among the eight most important infectious pathogens in hospitals (35). Nosocomial Klebsiella infections are caused mainly by K. pneumoniae, the most clinically important species of the genus, present as a saprophyte in the human mouth, the nasopharynx, and the intestinal tract (35). In Taiwan, the high prevalence of K. pneumoniae has also been observed. For example, a survey taken during 1991 to 2003 at a university hospital in Taiwan indicates that K. pneumoniae ranked second among gram-negative bacteria causing nosocomial infections, only after Escherichia coli (22). Recently, Klebsiella infection-induced liver abscess was first reported in Taiwan, and K. pneumoniae has surpassed E. coli, the historically predominant causative agent of hepatic abscesses, as the number one isolate from patients with this disease (56, 57). Since the initial description of extended-spectrum β-lactamase production by K. pneumoniae strains in 1983 (24), this organism's resistance to expanded-spectrum β-lactam antibiotics has emerged quickly (7). A similar situation was encountered in Taiwan (61), and it has caused increasingly serious problems in treating K. pneumoniae infections. Thus, a different approach, such as treatments with specific lytic bacteriophages, could be a possible alternative therapy (for a review, see references 3, 9, 25, 46, 47, and 48).
In this study, the first effort toward phage therapy for treating K. pneumoniae infections, we isolated a lytic bacteriophage (Kpp95) from hospital samples, using a clinical isolate of K. pneumoniae as the indicator cell. Characterization of this phage is presented here.
MATERIALS AND METHODS
Bacterial strains and cultivation.
Bacterial strains used in this study are listed in Table 1. All the strains were identified on the basis of microbiological methods and verified by using a VITEK system (BioMerieux Vitek Inc., Hazelwood, MO). Luria-Bertani (LB) agar and LB broth (36) were the media used for bacterial growth at appropriate temperatures with aeration. Antibiotics used were ampicillin (50 μg/ml) and kanamycin (50 μg/ml).
TABLE 1.
Bacterial strains used in this study
| Strain(s) | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Acinetobacter baumannii | ||
| Ab1-Ab10 | Imir Merr Ampr | This studyb |
| Enterobacter agglomerans | ||
| EaJ0-EaJ9 | EaJ0 (ESBL strains), Ampr | This studyb |
| Enterobacter cloacae | ||
| Ec1-Ec12 | ESBL strains, Ampr | This studyb |
| Escherichia coli | ||
| DH5α | endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA relA1 φ80d lacZΔ M15Δ (lacZYA-argF)U169 | 18 |
| E1-E13 | Ampr | This studyc |
| Klebsiella oxycota | ||
| KoJ1-KoJ8 | Ampr | This studyb |
| KoC1-KoC6 | Ampr | This studyc |
| Klebsiella pneumoniae | ||
| 10693 | Phage host, Amps | ATCC 23357 |
| Kp1-Kp108d | ESBL strains, Ampr | This studyc |
| Proteus mirabilis | ||
| Pm1, Pm3, Pm18, Pm40, Pm51, Pm59, Pm69, Pm70, Pm72, Pm73, Pm84, Pm97, Pm103, Pm111 | ESBL strains, Ampr | 59 |
| Pseudomonas aeruginosa | ||
| 27853 | Amps | 38 |
| Pa1-Pa6 | Imir Merr Ampr | This studyb |
| Serratia marcescens | ||
| Sm1-Sm4, Sm8 | ESBL strains, Ampr | 58 |
Amp, ampicillin; Imi, imipenem; Mer, meropenem; r, resistant; s, susceptible; ESBL, extended-spectrum β-lactamase.
Strains isolated in Jen-Ai Hospital, Taichung, Taiwan, Republic of China.
Strains isolated in the Hospital of China Medical University, Taiwan, Republic of China.
All of these strains were Ampr, but only 25 of them were ESBL strains.
Spot test and plaque assay.
To detect the presence of phage in a sample and the phage sensitivity of a bacterium, spot tests were carried out as described previously (11), except that LB broth and LB agar plates were used. To determine the phage titers, double-layered bioassays were performed on LB agar plates with the top and bottom layers containing 0.75% and 1.5% agar, respectively. One-tenth of a milliliter each of a phage suspension after serial dilutions and 100 μl of K. pneumoniae 10693 from an overnight culture were mixed with 3 ml of molten soft agar and poured onto the bottom solidified agar (12 ml). The numbers of plaques were counted after incubating the plates overnight. The same method was used to confirm phage susceptibility with the cells of different bacteria as the indicator hosts.
Propagation and purification of phage.
To propagate the phage, overnight cultures of K. pneumoniae 10693 were diluted 20-fold into 125-ml flasks containing 20 ml of the LB broth. When the cultures reached mid-exponential phase, the phage particles were added at a multiplicity of infection (MOI) of 20 and further incubated until ca. 12 h postinfection. Crude phage suspensions were prepared by centrifugation (10,000 × g, 15 min, at 4°C) of the cultures to remove the cells and by passing the supernatants through a membrane filter (0.45-μm-pore size). Phage particles were purified by banding in ultracentrifugation as previously described (23).
One-step growth curve.
A culture (10 ml in a 125-ml flask) of K. pneumoniae 10693 grown to mid-exponential phase was harvested by centrifugation and resuspended in fresh LB broth (ca. 1 × 109 CFU/ml). Phage was added at an MOI of 0.003 and allowed to adsorb for 5 min at 4°C. The mixture was then centrifuged, and the pelleted cells were resuspended in 10 ml of LB, followed by incubation at 37°C. Samples were taken at 5-min intervals and immediately titered by the double-layered agar plate method (34). The first set of samples were immediately diluted and plated for phage titration. A second set of samples was treated with chloroform (1% final concentration) to release intracellular phage for determining the eclipse period before phage titration. Experiments were repeated three times with duplicate samples.
Isolation of phage DNA.
Phage DNA was purified either by a Wizard Lambda DNA Preps DNA purification system (Promega, Madison, WI) or by phenol extraction of concentrated phage particles. In the latter case, the phage lysate was incubated for 30 min at 37°C in the presence of 20% polyethylene glycol 8000-2.5 M NaCl. The mixture was then incubated on ice for 60 min followed by centrifugation in 15-ml Corex tubes (Corning, Corning, NY) at 12,000 × g for 5 min. The pellet was suspended in 400 μl of 40 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 10 mM MgSO4. The mixture was clarified by centrifugation in a microcentrifuge for 2 min. The supernatant was extracted twice with chloroform. Phage DNA was then released by gentle mixing in an equal volume of Tris-HCl (40 mM, pH 7.9)-buffered phenol for 5 min. After centrifugation for 5 min, the upper layer was extracted again with phenol and then with chloroform. DNA was precipitated by the addition of 1 ml of 95% ethanol and 50 μl of 3 M sodium acetate (pH 5.2), collected by centrifugation, rinsed with 300 μl of 70% ethanol, allowed to dry, and gently suspended in 50 μl of 10 mM Tris-HCl (pH 8.0) containing 0.1 mM EDTA.
Restriction endonucleases were purchased from Takara Shuzou (Shiga, Japan), Gibco BRL (Grand Island, NY), or Roche Molecular Biochemicals (Indianapolis, IN). Digestion of the phage DNA was performed according to the instructions provided by the suppliers. The samples were electrophoresed either in 0.8% agarose (0.5× Tris-borate-EDTA buffer [TBE]) at 50V for 1 h or in 1% agarose (0.5× TBE buffer) at 50 V for 3 to 4 h.
PCR amplification.
To amplify the internal fragment of Kpp95 gene 23, we used primers P23F (5′-TGTTATAGGTATGGTACGACGTGCTAT-3′) and P23R (5′-TGAAGTTACCTTCACCACGACCGG-3′), which were previously used for PCR amplification of a 789-bp fragment internal to the gene 23 of various T4-type phages (49). PCR mixtures (10 μl) consisted of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.5 μM each of the primers, 0.16 mM deoxynucleoside triphosphates (Gibco BRL), 0.5 U of Taq polymerase (Gibco BRL), and 1 μl of phage lysate (104 to 107 PFU). Reactions were performed at 95°C for 2 min, followed by 30 cycles of 95°C for 60 s, 53°C for 60 s, and 72°C for 60 s. The sizes of the amplified fragments were checked with agarose gel (1.5% in 0.5× TBE buffer) electrophoresis run at 50 V, followed by ethidium bromide staining.
Pulsed-field gel electrophoresis.
Pulsed-field gel electrophoresis (PFGE) was performed in a CHEF-DR II PFGE system (Bio-Rad Laboratories, Hercules, CA) as described previously (52). The 1% gels were made with Seakem Gold agarose (Rockland, ME) in 0.5× TBE buffer. The conditions for electrophoresis were 0.5× TBE running buffer, 1-s initial switch time, 40-s final switch time, 200 V, 20-h running time, and a buffer temperature of 14°C. Lambda DNA concatemers, HindIII-digested DNA, and Saccharomyces cerevisiae YPH80 chromosomes (New England Biolabs, Beverly, MA) were used as the molecular size markers. The gels were stained with ethidium bromide and visualized on a UV box.
SDS-PAGE and protein analysis.
For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation, sample containing the Kpp95 was mixed with the sample buffer and then heated in a boiling water bath for 3 min, as described previously (27), and electrophoresed in an SDS-polyacrylamide gel (12%) by the method of Laemmli (25). Protein bands were visualized by staining the gels with Coomassie brilliant blue. For determination of the N-terminal amino acid (aa) sequence, the Kpp95 was separated by SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane, and then the protein was eluted from the membrane and subjected to Edman degradation in an Applied Biosystems model 476A protein sequencer. Protein concentration was determined as previously described (26).
DNA cloning and sequencing.
The Kpp95 DNA isolated from the phage particles was digested with SspI, and the resultant fragments were blunt ended and cloned into the E. coli vector pOK12 (54). Direct sequencing of both strands of the PCR products or the inserts was performed by dideoxy chain termination with an Applied Biosystems sequencer (ABI 377) (41) (Table 2).
TABLE 2.
Proteins/protein segments deduced from sequenced Kpp95 DNA fragments and their identities to homologues of T4-type phages
| Fragment (gene product)a | Corresponding aa positions at T4 homologueb | T4 protein | Identities shared (%) with corresponding proteins from T4-type phages | Accession no. |
|---|---|---|---|---|
| 906 (gp17) | 97-399 (610) | Terminase (central region) | RB69 (89); T4, RB32 and JS98 (87) | DQ845391 |
| 3788 (gp17) | 414-610 (610) | Terminase (C terminus) | RB69 (83); RB32 and T4 (81); JS98 (79) | AY538772 |
| 3788 (gp18) | 1-656 (659) | Tail sheath | JS98 (76); RB69, RB32 and T4 (71) | AY538772 |
| 3788 (gp19) | 1-163 (163) | Tail tube | JS98 (82); RB43 (73); RB69 and 42 (69) | AY538772 |
| 3788 (gp20) | 1-156 (524) | Head portal | JS98 (69); RB69 and RB32 (65); T4 (64) | AY538772 |
| 789 (gp23)c | 98-360 (521) | Major capsid | JS98, T4 and JS9 (83); T6, RB69, RB32, AR1, MVSS and JSD1 (82) | DQ845392 |
| 858 (gp34) | 1,062-1,288 (1,289) | Proximal tail fiber | RB43 (40); JS98 (31); RB32 (29); T4 (28) | DQ845394 |
| 333 (gp37) | 96-165 (1,026) | Long tail fiber | K3 (49); T4 (47); T5 (46); JS98 and Ac3 (35) | DQ845393 |
| 1031 (gp41) | 1-142 (475) | Primase-helicase | RB69, RB32 and T4 (69); JS98 (65) | DQ845390 |
| 678 (gp wac) | 76-259 (487) | Whisker antigen | RB43 (43); T2 (38); T4 and K3 (37); JS98 and AR1 (36) | DQ845389 |
gp18 and gp19 are full-length proteins, whereas the others are segments.
Numbers in parentheses represent the full length of the T4 protein.
A PCR fragment.
Bioinformatics.
Amino acid sequence identity was searched by using BLAST and Blastp (http://www.ncbi.nlm.nih.gov). Sequences were aligned by CLUSTALW (http://www.ebi.ac.uk/clustalw), and phylogenetic trees based on the neighbor-joining method were constructed by using PHYLIP (version 3.6). A bootstrap confidence value was assigned for each branch of the tree using the SEQBOOT program of PHYLIP (1,000 replicates).
Nucleotide sequence accession numbers.
The Kpp95 sequences described here have been deposited in GenBank with the accession numbers DQ845389 to DQ845394 and AY538772 as listed in Table 2.
RESULTS AND DISCUSSION
Kpp95 is one of the phages isolated on K. pneumoniae.
During this study, 254 hospital samples, including patient specimens, catheter washings, and wastewater from drainages, were collected. These samples were screened separately for the presence of phage by spot testing using K. pneumoniae 6 (a clinical strain) and K. pneumoniae 10693 as indicator host cells. Twelve phages were isolated after three consecutive single-plaque isolations and designated as Kpp2, -3, -4, -5, -6, -7, -10, -11, -30, -42, -50, and -95. They were indistinguishable based on preliminary results of study of phage properties, including morphology, EcoRV restriction patterns, and genome sizes estimated by PFGE (data not shown except for that of Kpp95 which is described below). Kpp95, capable of rapid lysis of host cells yielding high titers of progeny (3 × 1010 to 5 × 1010 PFU/ml), was studied further.
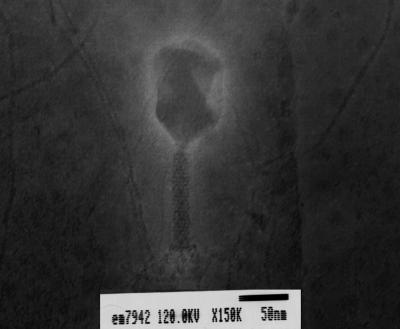
Electron microscopy showed that Kpp95 had a moderately elongated icosahedral head (ca. 101 nm long and 72 nm in diameter) connected to a sheathed tail (ca. 100 nm long and 13 nm wide) (Fig. 1). The distal end of the tail contained structural elements similar to those of the base plate, and a collar was localized faintly at the head-tail junction (Fig. 1). This virion morphology was similar to that of the T-even phages (2).
FIG. 1.
Electron micrograph of bacteriophage Kpp95. The sample was stained with 2% uranyl acetate.
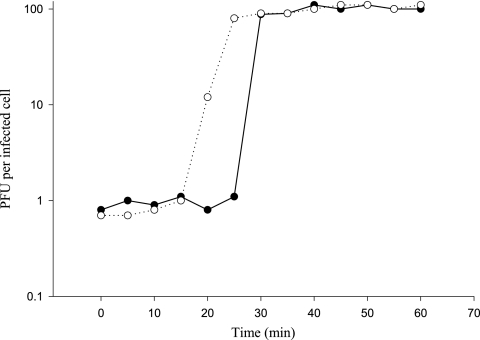
The one-step growth curve of Kpp95 was measured as described in Materials and Methods on K. pneumoniae strain 10693. As shown in Fig. 2, eclipse and latent periods of 20 to 25 and 30 min, respectively, were observed, indicating that Kpp95 requires a short growth period of about 5 to 10 min. The estimated burst size was 100 PFU per infected cell (Fig. 2). These values fit into the ranges observed for T-even group phages (1).
FIG. 2.
One-step growth curve of bacteriophage Kpp95 on K. pneumoniae strain 10693 in chloroform-treated culture (○) and in untreated cultures (•).
Kpp95 has a double-stranded DNA genome that appears to have base modifications.
T-even genomes contain hydroxyl-methylcytosine in place of cytosine, and these residues are generally glucosylated, rendering the DNA resistant to digestion by some restriction enzymes, which provides additional protection against host restriction systems (5, 28). In contrast, the genomes of the pseudo-T-even phages can be cut by restriction enzymes that are unable to digest T-even DNA, suggesting that only limited nucleotide modifications must be present in the pseudo-T-even genomes (32). We were interested in understanding whether the Kpp95 DNA could be cut by restriction endonucleases to give distinct fragments from which we might be able to sum up the total lengths for estimation of the genome size. To achieve this, DNA was prepared from Kpp95 phage particles and separately digested with 16 restriction endonucleases, including AseI, AvaI, BamHI, DraI, EcoRI, EcoRV, HaeIII, HincII, HindIII, KpnI, NaeI, PstI, SacII, Sau3A1, SmaI, and SspI. The DNA could be cut only by AseI, DraI, EcoRV, NdeI, and SspI into distinct bands, as visualized with an agarose gel after electrophoresis and staining with ethidium bromide (data not shown), but was refractory to digestion with the other 11 enzymes tested. Although the bands were not resolved well enough for estimation of the total lengths, susceptibility to restriction digestion indicated that the Kpp95 genome is a double-stranded DNA molecule. Notably, each of the five competent enzymes has a cleavage site flanked by either A and T (DraI, EcoRV, NdeI, and SspI) or T (AseI), suggesting that the Kpp95 genomic DNA contains a modified derivative(s) of G and/or C, rendering the DNA resistant to enzymes with cleavage sites flanked by G or C. These findings suggest that base modifications similar to the cases in T-type phages occur in Kpp95.
In parallel experiments, the restriction enzymes EcoRI, HincII, HindIII, and PstI that could not cut the Kpp95 genome were found capable of cutting the chromosomal DNA prepared from K. pneumoniae 10693 (data not shown), indicating that the same base modification is not found in the host cell. In addition, randomly cloned fragments of Kpp95 genome prepared from E. coli (see below) were susceptible to digestion with these restriction enzymes. These observations together indicate that the modification system is encoded in Kpp95.
Kpp95 genome is ca. 175 kb in length.
Since our efforts to estimate the Kpp95 genome size by summing the restriction fragment lengths were unsuccessful, we adopted pulsed-field gel electrophoresis to mobilize the intact genomic DNA prepared from the phage particles. As shown in Fig. 3, the Kpp95 genome migrated to a distance corresponding to a size of 170 to 180 kb. T4 packages its genome by a head-filling mechanism (45); and with similar head sizes, pseudo-T-even phages have genomes with sizes comparable to that of T4 (49). Thus, the Kpp95 has a genome size falling within the range of those of T4 (168,903 bp) and the pseudo-T-even phages (31, 49).
FIG. 3.
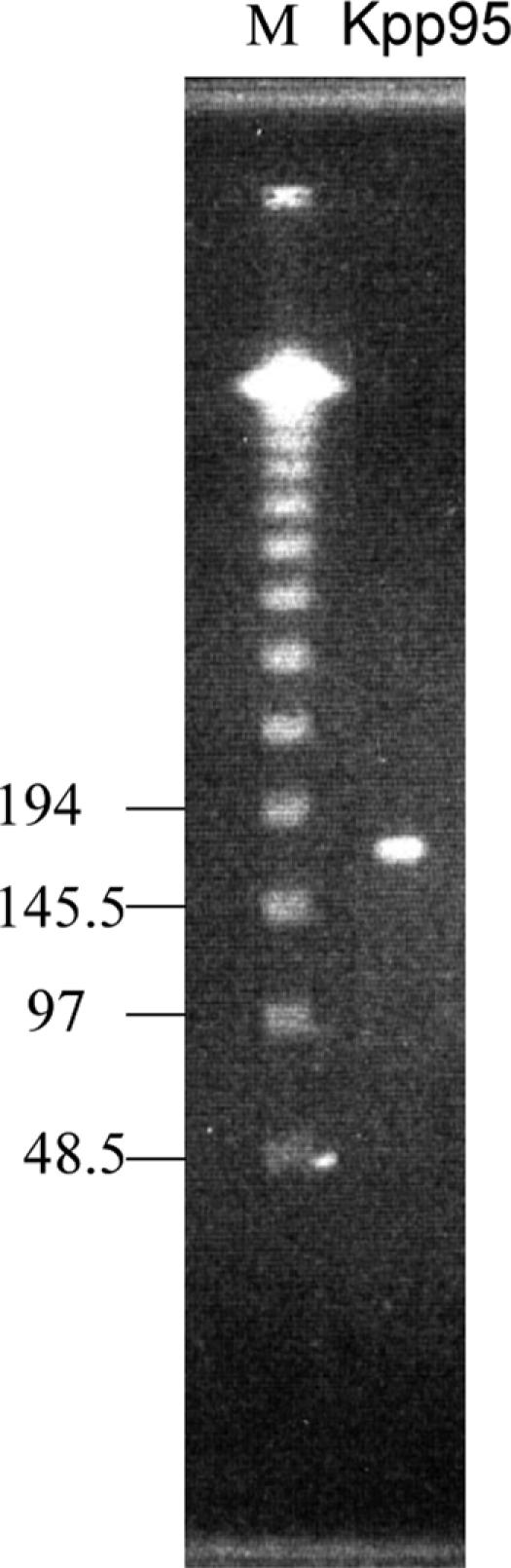
PFGE of the Kpp95 genome. Electrophoresis was performed with 1% agarose gel at 14°C and 200 V, with an initial time of 1 s, a final time of 40 s, and a running time of 20 h. Lane M contains molecular size markers (Lambda DNA ladder).
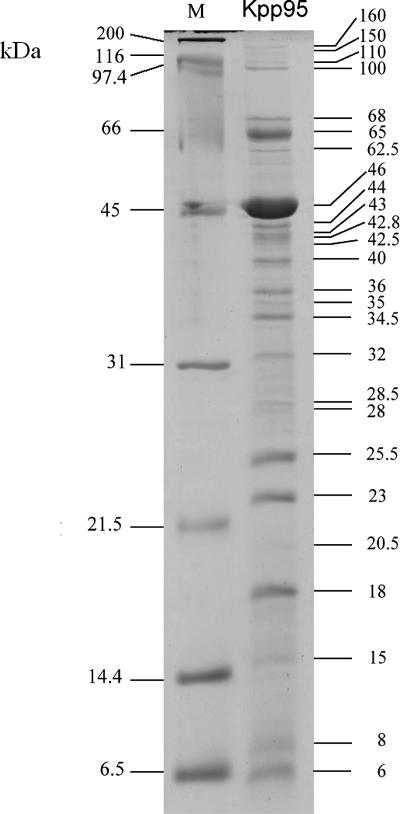
At least 26 virion proteins of Kpp95 can be visualized by SDS-PAGE.
To analyze the virion structural proteins, the Kpp95 phage particles were purified by ultracentrifugation, treated by boiling with the cracking buffer, and separated by SDS-PAGE (Fig. 4). At least 26 distinct protein bands, with molecular masses ranging from 6 to 200 kDa, were visualized after the gels were stained with Coomassie brilliant blue. The most abundant one in the gel was a 46-kDa protein, most likely the major coat protein of Kpp95. The sequence of the N-terminal 15 aa chemically determined for this protein, AEIGGDHGYDAQNIA, was 14/15 identical to aa 67 to 81 of the SV14 and RB69 gp23 proteins and 13/15 identical to aa 66 to 80 of the T-even (T2 and T6) and AR1 gp23 proteins (32, 49, 53, 60). In T4, during phage maturation, the N-terminal 65 aa residues of gp23 (521 residues) are cleaved off to give rise to gp23* (residues 66 to 521) (44). Due to the high abundance of the protein and the high degree of identity to the mature N terminus of the T4 gp23, it is reasonable to conclude that this 46-kDa protein is the mature major coat protein of Kpp95 that has been N-terminally processed to cleave off a length similar to that of T4 gp23.
FIG. 4.
SDS-PAGE of Kpp95 coat proteins. Lane M contains broad-range protein molecular mass markers.
Random genome sequencing indicates Kpp95 is similar to T-even phages.
In order to understand its similarity to other phages, the Kpp95 genome was digested with SspI, and the resultant fragments were blunt ended and cloned into pOK12. Six of the recombinant plasmids were subjected to nucleotide sequence determination of the inserts. Analysis revealed 906-, 3,788-, 858-, 333-, 1,031-, and 678-bp sequences. The G+C contents of these regions were around 40%, ranging from 38 to 42%, which deviated from that (55.5%) of the host genome (6). Notably, 2, 9, 3, 2, 2, and 2 recognition sequences for restriction endonucleases EcoRI, HincII, HindIII, KpnI, NaeI, and PstI, respectively, that could not cut the Kpp95 DNA were found in these sequenced fragments.
The sequences of the above-cited fragments showed low degrees of similarity at the nucleotide level to the entries in the database. However, alignment of the aa sequences deduced from these DNA sequences revealed that varied degrees of identity were shared with proteins from T4-type phages. Table 2 lists (i) the products of the genes residing in the fragments, (ii) the aa positions corresponding to those of T4, (iii) the names of homologous proteins in T4, and (iv) at least four homologues from T4-type phages that shared the highest degrees of identity (36 to 89%) with the deduced Kpp95 proteins or protein segments. The 906-bp fragment encoded a region homologous to aa 97 to 399 of T4 gp17 (terminase, a packaging protein). The region homologous to aa 414 to 610 of the T4 gp17 was found in an upstream region of the 3,788-bp fragment. The remaining part of the 3,788-bp fragment encoded the complete gp18 (tail sheath protein) and gp19 (tail tube protein) and the N terminus of gp20 (head portal protein). The 858-, 333-, 1,031-, and 678-bp DNA fragments encoded protein segments homologous to the corresponding regions of T4 gp34 (proximal tail fiber protein), gp37 (long tail fiber protein), gp41 (primase-helicase), and gp wac (whisker antigen), respectively. Table 2 also shows the identities (82 to 83%) shared between the central region of the major capsid protein gp23 from T4-type phages (corresponding to aa 98 to 360 of T4 gp23) and that from Kpp95 revealed by sequencing a PCR fragment (263 aa) amplified on the Kpp95 genome by using a T4 gp23-specific degenerate primer pair (see below).
It is worth noting that (i) the sequences from JS98, RB69, RB43, RB32, and T4 were always the ones sharing the highest degrees of identity with the Kpp95 proteins and that (ii) while over 64 to 89% identities were shared among gp17, gp18, gp19, gp20, gp23, gp41, and the respective corresponding proteins, lower than 50% identities were shared among the gp34, gp37, and whisker protein homologues (Table 2).
Phylogenetic analysis of gp18, gp19, and gp23 indicated Kpp95 was most closely related to coliphage JS98, which classified it as a pseudo-T-even subgroup member.
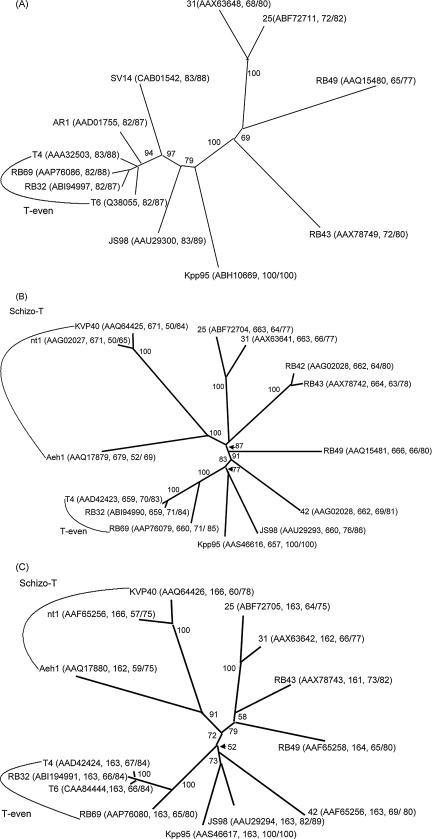
Based on relatedness of the sequences of three major virion structural proteins, the tail sheath protein (gp18), the tail tube protein (gp19), and the major capsid protein (gp23), the T4-type phages can be divided into four subgroups with increasing divergence from T4: the T-even, the pseudo-T-even, the schizo-T-even, and the exo-T-even phages (13). Although the phylogenetic trees obtained for all of the three virion proteins are similar, the ones based on gp23 are most commonly used for relatedness analysis (13, 15, 49). For the analyses, alignments were performed among the amino acid sequences encoded by the internal fragments, each of which was flanked by two regions of consensus among the gp23 of T4-type phages. In the T4 gp23 proteins, the internal region encompasses aa 95 to 375, and therefore, using the primer pair corresponding to aa 95 to 103 and 368 to 375 of the T4 gp23, the central portion of gene 23 (ca. 850 bp) of various T4-type phages can be amplified, and an internal region of 188 aa (aa 115 to 302, corresponding to the T4 gp23) was used for phylogenetic analysis (13, 49). In this study, we used the degenerated primers described in Materials and Methods for amplification, with the Kpp95 genome as the template, and an amplicon of 789 bp (encoding 263 aa) was obtained. To discern the relatedness, the aa sequence deduced from this Kpp95 fragment (instead of the 188-aa region, see below) was aligned with the corresponding region of gp23 from T4-type phages, which was available from the database, had a deposited length of at least 263 aa, and shared over 65% identity with that of the Kpp95 gp23. Eleven entries were found that met these criteria and were used for the construction of a phylogenetic tree as described in Materials and Methods. As shown in Fig. 5A, the analysis indicated that Kpp95 is most closely related to the coliphage JS98 (sharing the highest identity, 83%, with the Kpp95 gp23 region) belonging to the pseudo-T-even subgroup (Fig. 5A) (12, 13, 49).
FIG. 5.
Phylogenetic trees based on the aa sequence of gp23 (A), gp18 (B), and gp19 (C) from T4-type phages. The numbers at branching points are bootstrap values based on 1,000 replicates. Parentheses contain the GenBank accession numbers, followed by the lengths (aa) of the protein/protein segment (except that each of the gp23 segments was 263 aa in length) and then identity/similarity. The sequences include (i) T-even phages T4, T6, RB32, and RB69 infecting E. coli; (ii) pseudo-T-even phages 25 and 31 infecting Aeromonas salmonicida, phage 42 of Burkholderia cepacia, and coliphages AR1, JS98, RB42, RB43, RB49, and SV14; and (iii) schizo-T-even phage Aeh1 of Aeromonas hydrophila, KVP40 of Vibrio cholerae, and nt1 of Vibrio natriegens.
Full-length sequences of gp18 and gp19 from some T4-type phages were also available in the database. Among them, 13 each of gp18 and gp19 sharing the highest identities with that of the Kpp95 sequence were aligned for phylogenetic analyses. As shown in Fig. 5B and C, they showed 50 to 76% and 57 to 82% identities to the Kpp95 gp18 and gp19 sequences, respectively, with the highest degree of identity shared with the proteins from JS98 (76% for gp18 and 82% for gp19). The phylogenetic analyses based on the gp18 and gp19 sequences indicated that JS98 was still the phage most closely related to Kpp95.
During these analyses, we also tried to use the internal 188-aa sequences of gp23 for tree construction as described previously (13, 49). However, the relatedness found was closest to that of Burkholderia cepacia phage 42 (data not shown), which did not even appear in the tree when the sequences of 263 aa were based for gp23 analysis (Fig. 5A). Furthermore, when the full-length sequences were used for analysis of gp18 and gp19, the phage 42 sequences were not as closely related as those of JS98. Since the three trees consistently favored relatedness to JS98, it appears that the 263-aa sequences of gp23 can provide a better basis than the 188-aa internal regions for the relatedness analysis of T4-type phages.
Kpp95 has an extended host range.
To evaluate host susceptibility to Kpp95, the bacterial lawns of 107 K. pneumoniae strains, except for the indicator host strain 10693, were subjected separately to spot testing (Table 1). Completely clean clearing zones were formed on 40 (47%) of the strains, 25 (23.2%) strains gave turbid clearing zones, and 43 (39.8%) strains exhibited resistance to the phage. In plaque assays, the degrees of plaque clarity were consistent with those of the clearing zones.
Susceptibility to Kpp95 was also tested with 86 clinical strains representing eight bacterial species other than K. pneumoniae (Table 1). Results (the number of sensitive strains over the total strains tested) indicated that Enterobacter agglomerans (7/10), Klebsiella oxytoca (14/14), and Serratia marcescens (5/5) were susceptible to Kpp95. All strains shown to be positive by spot test gave clear plaques in plaque assays, except for S. marcescens, on which turbid plaques were formed. In these positive cases, plating efficiencies were similar when the same batches of the phage lysate were used for different host strains. Acinetobacter baumannii (10), Enterobacter cloacae (12), E. coli (14), Proteus mirabilis (14), and Pseudomonas aeruginosa (7), with the total numbers of strains tested in parentheses, were found to be resistant to Kpp95.
T4 infects only E. coli and the closely related Shigella sp., with the adsorption specificity being determined by the tip-of-tail fibers (gp37), which bind to receptors on the surface of the bacteria, and its host range is expanded by duplications of a small region of the tail fiber adhesin (50). Several other bacteriophages exhibiting an intrinsic broad host range have been reported, such as LG1 (a Myoviridae coliphage with 49.5-kb genomic DNA), the T4-like coliphage AR1, and the T4-like vibriophage KVP40. LG1 and AR1 can infect many serotypes of E. coli and enterobacteria, including Proteus mirabilis, Shigella dysenteriae, and two Salmonellas trains (16). KVP40 can infect eight vibrio species, including V. parahaemolyticus, V. cholerae, the nonpathogenic V. natriegens, and Photobacterium leiognathi (30). However, the mechanisms involved in these phages' abilities to infect different hosts have not been elucidated. To determine the sequence which might be involved in Kpp95 adsorption, we tried to obtain Kpp95 gene 37 by PCR amplification with primers specific to T4 gene 37 (37RV-1F [5′-GTTCTGGTAATTTTGCTAAC-3′] and 37RV-1R [5′-AACAGCTAACTTTGGATATG-3′], FR86 [5′-GCTTCAAGTACTGACTTAGG-3′], and FR89 [5′-ACAGTGATAGTATGACCATGTGATCC-3′]) (37, 49). However, the reactions failed to give a PCR product. This result was consistent with the low identity (47%) revealed by sequencing the fragment of g37 (Table 2) and indicated that the two phages have very divergent tail fibers.
Haggard-Ljungquist et al. (1992) have summarized the conditions that may cause a phage to alter or extend its host range, as follows (17). First, point mutations in the tail fiber genes can alter the host range. This might have evolved to counteract host range mutants of the bacterial hosts. Second, a new host range may be acquired by obtaining a portion of a tail fiber gene from another phage or from a gene left in the bacterial chromosome by another phage. This has been evidenced by (i) similarities in the tail fiber genes of such unrelated phages as P1, P2, Mu, λ, and T4, and (ii) by segments similar to genes 36 and 37 (tail fiber gene) of T2, and a cryptic gene (CRF86) similar to the tail fiber assembly gene have been detected in the E. coli chromosome (39). Third, phages Mu and P1 are capable of phase variations that alternate between two mutually exclusive sets of tail fiber genes, rendering the phages able to infect different hosts. Fourth, some phages such as λ and T5 may simultaneously use two separate systems of tail fibers, each recognizing a different receptor (8). In λ, the J gene product forming the single terminal tail fiber is used to recognize its normal receptor, LamB (38). In addition, the laboratory strain of λ carries defective genes stf and tfa, which encode the side-tail fibers. It is believed that reversion of the 1-bp deletion in stf by compensatory DNA replication can enable the phage to use the outer membrane protein C (OmpC) as a receptor (33). Phage T5 has two sets of tail fibers, the L-shaped tail fibers (which bind to lipopolysaccharide) and a single straight terminal tail fiber (which binds to FhuA receptor protein) (8, 19, 20, 40). Recently, a similar finding has been extended to the T7-like coliphage φK1-5, which has two different tail fiber proteins capable of infecting strains with a K1 or K5 polysaccharide capsule (42). Fifth, the same tail fiber of a phage may have the ability to recognize two separate receptors, e.g., T4 can use its tail fibers to recognize OmpC or the lipopolysaccharide as a receptor with equivalent efficiency (33). This last possibility may be the case for Kpp95, because (i) although the presence of two sets of tail fiber has been observed in some of the phages mentioned above, a similar situation has not been reported for T4-type phages, and (ii) the tail fiber adhesins of T-even phages are hotspots of lateral gene transfer not only between T4-type phages but also between different groups of morphologically distinct coliphages (17), giving hypervariable properties to the gp37 of T-even phages (reviewed in reference 21) which may also be exploited by Kpp95 for receptor recognition and adaptation to extended host range.
The emergence of pathogenic bacteria resistant to multiple drugs has posed a growing threat, and exploring alternative approaches such as phage therapy is a worthwhile task. While efforts are being increasingly directed along this line in North America and western Europe, numerous cases of successfully treated bacterial infections have long been known in Poland, Georgia, and the former Soviet Union, about which voluminous reports have been published, mostly in languages other than English, and some of these cases involved treatment of K. pneumoniae infections with phages (for a review, see references 3, 9, 29, 46, 47, and 48). In phage therapy, one of the problems that remains to be solved is the development of phage resistance by the bacterial hosts. Therefore, it is generally accepted that the isolation of host range mutant phages and the use of cocktails containing several phages in one preparation should be necessary to reduce the probability of resistance development and to cover a breadth of host ranges (31, 40). The lytic phage Kpp95, able to cause rapid and clear lysis of many K. pneumoniae strains, is worthy of development into a component of a therapeutic cocktail, maybe in conjunction with K. pneumoniae phages, which have previously been characterized as virulent (4, 10, 14, 43, 51, 55), or with antibiotics. Furthermore, with a broad host range, a therapeutic agent based on Kpp95 has the potential to treat coinfections by E. agglomerans, K. oxytoca, and S. marcescens.
Acknowledgments
This work was supported by grant CMU-93-M-11 from China Medical University.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Abedon, S. T., P. Hyman, and C. Thomas. 2003. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl. Environ. Microbiol. 69:7499-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, H. W., and H. M. Krisch. 1997. A catalogue of T4-type bacteriophages. Arch. Virol. 142:2329-2345. [DOI] [PubMed] [Google Scholar]
- 3.Alisky, J., K. Iczkowski, A. Rapoport, and N. Troitsky. 1998. Bacteriophages show promise as antimicrobial agents. J. Infect. 36:5-15. [DOI] [PubMed] [Google Scholar]
- 4.Benedi, V. J., M. Regue, S. Alberti, S. Camprubi, and J. M. Tomas. 1991. Influence of environmental conditions on infection of Klebsiella pneumoniae by two different types of bacteriophages. Can. J. Microbiol. 37:270-275. [DOI] [PubMed] [Google Scholar]
- 5.Berkner, K. L., and W. R. Folk. 1977. EcoRI cleavage and methylation of DNAs containing modified pyrimidines in the recognition sequence. J. Biol. Chem. 252:3185-3193. [PubMed] [Google Scholar]
- 6.Bradbury, J. F. 1989. Genus V. Klebisella Trevisan, 105AL, p. 226. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 7.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V., K. Schaller, and H. Wolff. 1973. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323:87-97. [DOI] [PubMed] [Google Scholar]
- 9.Brussow, H. 2005. Phage therapy: the Escherichia coli experience. Microbiology 151:2133-2140. [DOI] [PubMed] [Google Scholar]
- 10.Camprubi, S., S. Merino, V. J. Benedi, and J. M. Tomas. 1991. Isolation and characterization of bacteriophage FC3-10 from Klebsiella spp. FEMS Microbiol. Lett. 67:291-297. [DOI] [PubMed] [Google Scholar]
- 11.Chang, H.-C., C.-R. Chen, J.-W. Lin, G.-H. Shen, K.-M. Chang, Y.-H. Tseng, and S.-F. Weng. 2005. Isolation and characterization of novel giant Stenotrophomonas maltophilia phage φSMA5. Appl. Environ. Microbiol. 71:1387-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chibani-Chennoufi, S., C. Canchaya, A. Bruttin, and H. Brussow. 2004. Comparative genomics of the T4-like Escherichia coli phage JS98: implications for the evolution of T4 phages. J. Bacteriol. 186:8276-8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desplats, C., and H. M. Krisch. 2003. The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154:259-267. [DOI] [PubMed] [Google Scholar]
- 14.Dietz, A., H. Kossel, and R. Hausmann. 1985. On the evolution of the terminal redundancies of Klebsiella phage no. 11 and of coliphages T3 and T7. J. Gen. Virol. 66:181-186. [DOI] [PubMed] [Google Scholar]
- 15.Filee, J., F. Tetart, C. A. Suttle, and H. M. Krisch. 2005. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. USA 102:12471-12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodridge, L., A. Gallaccio, and M. W. Griffiths. 2003. Morphological, host range, and genetic characterization of two coliphages. Appl. Environ. Microbiol. 69:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Heller, K., and V. Braun. 1979. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 139:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heller, K., and V. Braun. 1982. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 41:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henning, U., and S. Hashemolhosseini. 1994. Receptor recognition by T-even-type coliphages, p. 291-298. In J. D. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, DC.
- 22.Hsueh, P. R., W. H. Chen, and K. T. Luh. 2005. Relationships between antimicrobial use and antimicrobial resistance in gram-negative bacteria causing nosocomial infections from 1991-2003 at a university hospital in Taiwan. Int. J. Antimicrob. Agents 26:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, C. H., H. C. Wu, and Y. H. Tseng. 2002. Mutation in the Xanthomonas campestris xanA gene required for synthesis of xanthan and lipopolysaccharide drastically reduces the efficiency of bacteriophage phiL7 adsorption. Biochem. Biophys. Res. Commun. 291:338-343. [DOI] [PubMed] [Google Scholar]
- 24.Knothe, H., P. Shah, V. Krcmery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lin, N.-T., T.-J. Liu, T.-C. Lee, B.-Y. You, M.-H. Yang, F.-S. Wen, and Y.-H. Tseng. 1999. The adsorption protein genes of Xanthomonas campestris filamentous phages determining host specificity. J. Bacteriol. 181:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, T. J., B. Y. You, N. T. Lin, M. T. Yang, and Y. H. Tseng. 1998. Purification and expression of the gene III protein from filamentous phage phi Lf. Biochem. Biophys. Res. Commun. 242:113-117. [DOI] [PubMed] [Google Scholar]
- 28.Malygin, E. G., W. M. Lindstrom, Jr., V. V. Zinoviev, A. A. Evdokimov, S. L. Schlagman, N. O. Reich, and S. Hattman. 2003. Bacteriophage T4Dam DNA-(adenine-N6)-methyltransferase: evidence for two distinct stages of methylation under single turnover conditions. J. Biol. Chem. 278:41749-41755. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki, S., M. Rashel, J. Uchiyama, S. Sakurai, T. Ujihara, M. Kuroda, M. Ikeuchi, T. Tani, M. Fujieda, H. Wakiguchi, and S. Imai. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J. Infect. Chemother. 11:211-219. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki, S., S. Tanaka, T. Koga, and T. Kawata. 1992. A broad-host-range vibriophage, KVP40, isolated from sea water. Microbiol. Immunol. 36:93-97. [DOI] [PubMed] [Google Scholar]
- 31.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monod, C., F. Repoila, M. Kutateladze, F. Tetart, and H. M. Krisch. 1997. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J. Mol. Biol. 267:237-249. [DOI] [PubMed] [Google Scholar]
- 33.Montag, D., S. Hashemolhosseini, and U. Henning. 1990. Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J. Mol. Biol. 216:327-334. [DOI] [PubMed] [Google Scholar]
- 34.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Press, R., N. Glansdorff, P. Miner, de J. Vries, R. Kadner, and W. K. Maas. 1971. Isolation of transducing particles of phi-80 bacteriophage that carry different regions of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 68:795-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu, Y., P. Hyman, T. Harrah, and E. Goldberg. 2004. In vivo bypass of chaperone by extended coiled-coil motif in T4 tail fiber. J. Bacteriol. 186:8363-8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall-Hazelbauer, L., and M. Schwartz. 1973. Isolation of the bacteriophage lambda receptor from Escherichia coli. J. Bacteriol. 116:1436-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riede, I., M. L. Eschbach, and U. Henning. 1985. Presence of DNA, encoding parts of bacteriophage tail fiber genes, in the chromosome of Escherichia coli K-12. J. Bacteriol. 163:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saigo, K. 1978. Isolation of high-density mutants and identification of nonessential structural proteins in bacteriophage T5: dispensability of L-shaped tail fibers and a secondary major head protein. Virology 85:422-433.664210 [Google Scholar]
- 41.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Souza, K. A., H. S. Ginoza, and R. D. Haight. 1972. Isolation of a polyvalent bacteriophage for Escherichia coli, Klebsiella pneumoniae, and Aerobacter aerogenes. J. Virol. 9:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steven, A. C., A. C. Bauer, M. E. Bisher, F. A. Robey, and L. W. Black. 1991. The maturation-dependent conformational change of phage T4 capsid involves the translocation of specific epitopes between the inner and the outer capsid surfaces. J. Struct. Biol. 106:221-236. [DOI] [PubMed] [Google Scholar]
- 45.Streisinger, G., J. Emrich, and M. M. Stahl. 1967. Chromosome structure in phage T4, III. Terminal redundancy and length determination. Proc. Natl. Acad. Sci. USA 57:292-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulakvelidze, A., and E. Kutter. 2005. Bacteriophage therapy in humans, p. 381-436. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL.
- 48.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437-451. [DOI] [PubMed] [Google Scholar]
- 49.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tetart, F., F. Repoila, C. Monod, and H. M. Krisch. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 258:726-731. [DOI] [PubMed] [Google Scholar]
- 51.Tomas, J. M., and J. T. Jofre. 1985. Lipopolysaccharide-specific bacteriophage for Klebsiella pneumoniae C3. J. Bacteriol. 162:1276-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng, Y.-H., K.-T. Choy, C.-H. Hung, N.-T. Lin, J.-Y. Liu, C.-H. Lou, B.-Y. Yang, F.-S. Wen, S.-F. Weng, and J.-R. Wu. 1999. Chromosome map of Xanthomonas campestris pv. campestris 17 with locations of genes involved in xanthan gum synthesis and yellow pigmentation. J. Bacteriol. 181:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsugita, A., and R. Van den Broek. 1987. The amino acid sequence of crystalline sheets: a proteolytic fragment of the major head protein (gP23) of bacteriophage T4. Protein Seq. Data Anal. 1:99-102. [PubMed] [Google Scholar]
- 54.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 55.Vinodkumar, C. S., Neelagund, Y. F., and S. Kalsurmath. 2005. Bacteriophage in the treatment of experimental septicemic mice from a clinical isolate of multidrug resistant Klebsiella pneumoniae. J. Commun. Dis. 37:18-29. [PubMed] [Google Scholar]
- 56.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, J. H. Wang, S. R. Wann, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26:1434-1438. [DOI] [PubMed] [Google Scholar]
- 57.Wong, W. M., B. C. Wong, C. K. Hui, M. Ng, K. C. Lai, W. K. Tso, S. K. Lam, and C. L. Lai. 2002. Pyogenic liver abscess: retrospective analysis of 80 cases over a 10-year period. J. Gastroenterol. Hepatol. 17:1001-1007. [DOI] [PubMed] [Google Scholar]
- 58.Wu, L. T., M. F. Tsou, H. J. Wu, H. E. Chen, Y. C. Chuang, and W. L. Yu. 2004. Survey of CTX-M-3 extended-spectrum beta-lactamase (ESBL) among cefotaxime-resistant Serratia marcescens at a medical center in middle Taiwan. Diagn. Microbiol. Infect. Dis. 49:125-129. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L. T., H. J. Wu, J. G. Chung, Y. C. Chuang, K. C. Cheng, and W. L. Yu. 2006. Dissemination of Proteus mirabilis isolates harboring CTX-M-14 and CTX-M-3 beta-lactamases at 2 hospitals in Taiwan. Diagn. Microbiol. Infect. Dis. 9. 54:89-94. [DOI] [PubMed] [Google Scholar]
- 60.Yu, S.-L., K.-L. Ko, C.-S. Chen, Y.-C. Chang, and W.-J. Syu. 2000. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J. Bacteriol. 2000. 182:5962-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, W.-L., K.-C. Cheng, L.-T. Wu, M. A. Pfaller, P. L. Winokur, and R. N. Jones. 2004. Emergence of two Klebsiella pneumoniae isolates harboring plasmid-mediated CTX-M-15 β-lactamase in Taiwan. Antimicrob. Agents Chemother. 48:362-363. [DOI] [PMC free article] [PubMed] [Google Scholar]