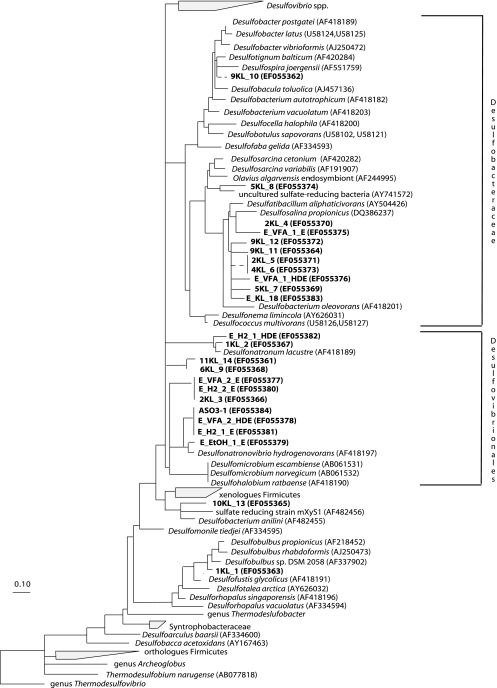
FIG. 2.
Consensus phylogenetic tree based on amino acid sequences of the dsrAB gene. The partial sequences determined in this study are in bold type. They were added to the tree using the QUICK_ADD parsimony tool and individually removed to avoid long branch attraction. Deletions and insertions were not included in the calculation. Branching orders that were not supported by all tree construction methods are shown as multiforcations. The band number (Fig. 1) is preceded by the lake number (Table 1) for sediment samples. For enrichment samples, the following code was used: E_H2/VFA/EtOH (ethanol [EtOH]) (electron donor)_1/2 (total Na+ [in molar concentration])_E/HDE/HDO/LDO (type of enrichment). E stands for enrichment obtained by regular transfers of an incubation that was originally inoculated with the sediment mixture sample. HDE are enrichments that were obtained by regular transfer of the highest positive dilution of the initial sediment incubation at that specific condition, whereas HDO and LDO are the first enrichments with the highest and the lowest positive dilutions, respectively, in which the original sediment sample was used as inoculum.