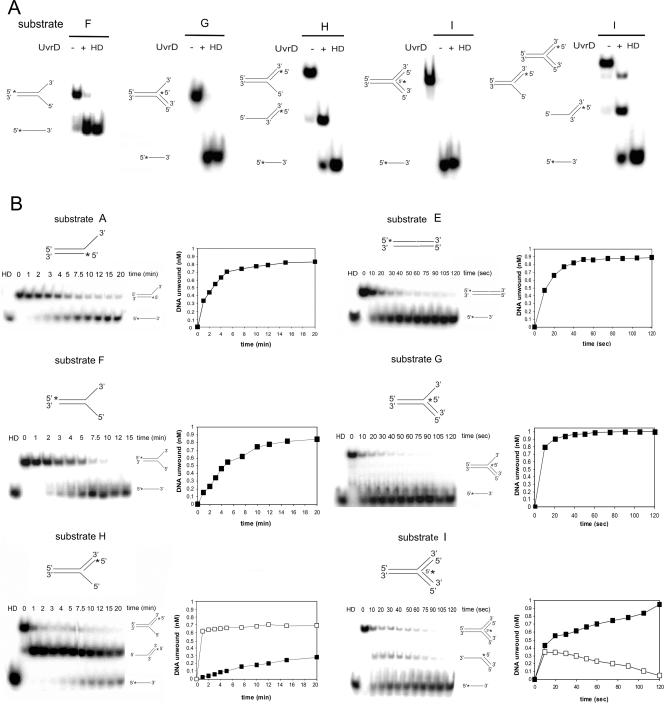
FIG. 5.
Unwinding of substrates with different structures by UvrD. (A) Substrates (1 nM) were incubated in the presence of 1 mM ATP, 5 mM MgCl2, and 200 nM UvrD at 37°C for 10 min. Reactions were analyzed by native PAGE, and PhosphorImager scans of the dried gels are shown. HD, heat denatured. The 32P label is indicated with asterisks. (B) Time course analysis of the unwinding of different substrates (A to I, as reported in Table 2). Each substrate (1 nM) was incubated in the presence of 1 mM ATP, 5 mM MgCl2, and 200 nM UvrD at 37°C for the indicated times. The data for the unwinding reactions were quantified and plotted; note that the units for the scales of the x axes are minutes for the left panels but seconds for the right panels. The amounts of labeled DNA unwound in time are represented as closed squares, while the intermediate structures are represented as open squares. The data reported are the averages for at least three independent experiments, and the gels are representative images.