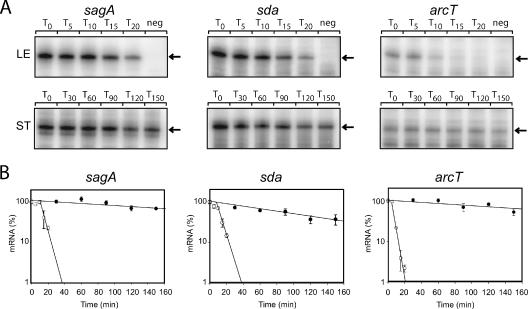
FIG. 1.
Decay rate of sagA, sda, and arcT transcripts in late exponential (LE) and stationary (ST) phases determined by RPAs. (A) RNA was extracted from MGAS315 at the times (minutes) indicated above each lane following the addition of rifampin to halt transcription. RPAs were performed as described previously (11) using 10 μg total RNA from exponential phase and 5 μg from stationary phase. A negative control for each probe (neg) consists of labeled probe without RNA. The arrow to the right of each gel indicates the expected size of the protected RNA probe following digestion with RNase T1. (B) The amount of each transcript in Fig. 1A was quantified using ImageQuant 5.0 software (Molecular Dynamics). The average intensity of each band was calculated for each time point. The y axes of the graphs represent the fraction (%) of each message remaining at the indicated time after addition of rifampin. Error bars represent the standard deviation of values obtained for each time point from two independent reactions. Regression analysis was performed using SigmaPlot 9.0 software (SPSS Inc.) and used to calculate the slope of each line. Open symbols, late exponential phase; closed symbols, stationary phase.