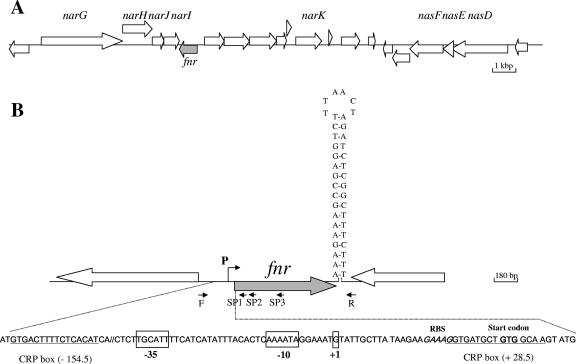
FIG. 2.
Genetic organization of the fnr chromosomal region of B. cereus F4430/73 and ATCC 14579. (A) Large arrows represent the open reading frames identified in strain ATCC 14579 (ERGO; Integrated Genomics, Inc.), and their orientation shows the transcriptional direction. Only the names of open reading frames encoding proteins with predicted metabolic functions are indicated. (B) The nucleotide sequences of the promoter region and the putative terminator of F4430/73 fnr are shown. Primers F and R were used for the amplification of the overall DNA of the fnr locus, and primers SP1, SP2, and SP3 were used to map the transcriptional start site. The transcriptional start site (+1) and the putative −35 and −10 motifs are boxed. Putative CRP boxes are underlined. RBS, ribosome-binding site.