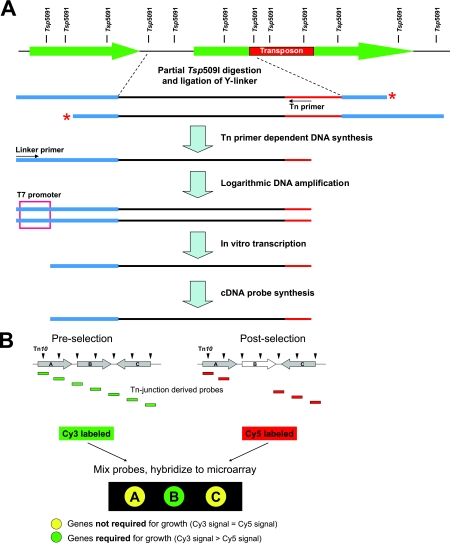
FIG. 1.
Schematic diagram of the TraSH screening strategy. (A) TraSH probe generation. Genomic DNA from a population containing transposon insertions (red) is partially digested with the four-base cutter Tsp509I and size fractionated; fragments are ligated to Y-shaped linker DNA (blue) which encodes an inactive T7 promoter but not the complementary strand. Ligation products are subjected to PCR amplification using a transposon-derived primer and a linker-derived primer which encodes a functional T7 promoter; logarithmic amplification of PCR products is dependent upon DNA synthesis from the transposon-derived primer, which generates the linker-derived primer binding site (extension of the linker, which would generate a binding site for the linker-derived primer, is prevented by a 3′ amino modification [red asterisk]). As a result, PCR products are generated only from sequences adjacent to transposon insertions. PCR products are transcribed in vitro from the T7 promoter, and amino-allyl-labeled cDNA probes are reverse transcribed from the RNA. (B) Microarray hybridization of TraSH probes. A population of mutants containing multiple transposon insertion mutations (indicated by arrowheads above genes A, B, and C) grown on rich medium (preselection) is subjected to growth under selective pressure (postselection). TraSH probes, PCR amplified from the mutant populations before and after growth selection and differentially labeled with fluorescent dyes, are cohybridized to microarrays. Clones harboring mutations in genes required for fitness under the selective pressure (gene B) are reduced or eliminated from the postselection population, resulting in reduced microarray hybridization signals compared to those of the parental preselection population. The figure was adapted from the study of Sassetti and Rubin (20).