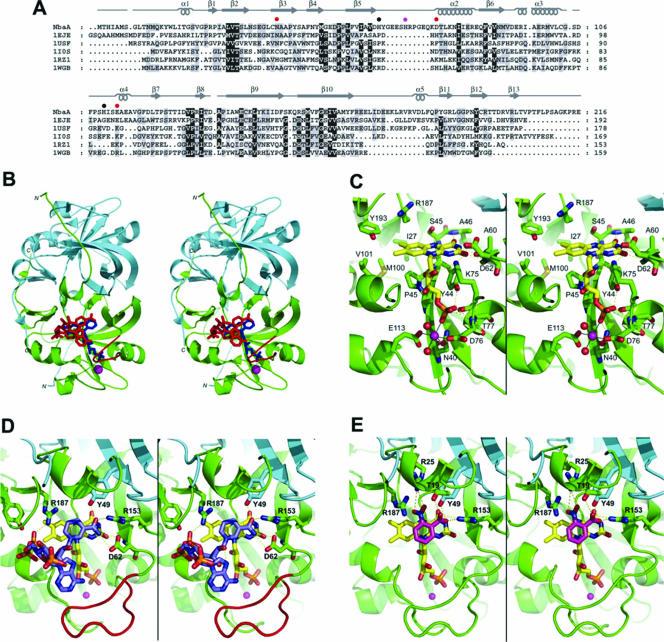
FIG. 4.
Modeled NbaA structure and interactions with FMN, NADPH, and 2-NBA. (A) Fold-recognition-based sequence alignment between NbaA and the currently structurally characterized members (identified here by their PDB accession numbers at http://www.rcsb.org/) of the NADH:FMN oxidoreductase-like structural family from the FMN-binding split-barrel fold: accession number 1EJE, Methanobacterium thermoautotrophicum FMN binding protein MTH152; accession number 1USF, Thermus thermophilus styrene monooxygenase; accession number 1IOS, Archeoglobus fulgidus ferric reductase; accession number 1RZ1, Bacillus thermoglucosidasius flavin reductase PheA2; accession number 1WGB, Thermus thermophilus probable flavoprotein. Secondary structure elements (α-helices and β-strands) of NbaA, corresponding to its three-dimensional homology model built based on this alignment, are indicated above its sequence. The NbaA residues individually mutagenized to alanine in this study are marked by closed circles that are color coded according to the activity of the corresponding mutants (red, inactive; magenta, residual activity; black, activity comparable to that of the wild type). (B) Stereo view displaying the structural model of the NbaA dimer. The trace of the NbaA main chain, rendered schematically by its secondary structure, is colored differently for the two monomers (green and cyan, respectively), with the 10-residue insertion in the β5-α2 loop relative to the other members of the fold family highlighted in red in one of the monomers. Ligands docked to NbaA are shown in only one of the two equivalent binding sites of the homodimer and are colored (blue, FMN; red, NADPH; magenta, Ni2+). (C) Stereo view zooming in the FMN binding site of NbaA. (D) Stereo view showing the modeled interactions of NADPH with the FMN-bound NbaA. (E) Stereo view detailing the putative binding mode of the 2-NBA substrate. In the latter three panels, the protein main chain is rendered as in B, select protein residues and side chains are shown explicitly, and the following color scheme is applied to the C atoms belonging to the various displayed molecules: yellow, FMN; light blue, NADPH (D); magenta, 2-NBA (E); green or cyan, protein. The other atom types are colored as follows: blue, N; red, O; yellow, S; orange, P. Hydrogen bonds are shown with yellow dashed lines, and metal ion hexacoordination is shown with black solid lines. The Ni2+ ion is represented as a magenta sphere, and its two coordinating water molecules are represented as red spheres in C.