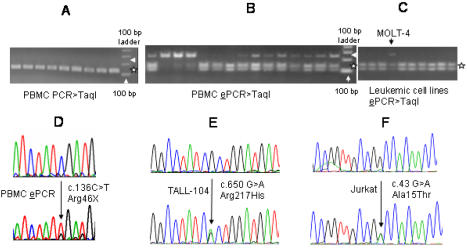
Figure 3. SDHB mutations in genomic DNAs (gDNAs).
A. TaqI RE digestion of PCR-amplified exon 2 shows no evidence of undigested mutant DNAs at 260 bp size (pointed by an arrow head). TaqI RE digestion products of the wild-type DNA co-migrate at sizes of 128 and 132 bps (denoted by a star). B. Gel electrophoresis of TaqI RE-digested (mutation) enrichment PCR (ePCR) products shows variable amounts of mutant gDNAs at 233 bp size (pointed by an arrow head), which are confirmed to be primarily composed of c.136C>T by sequencing (Table 1). TaqI digestion of wild-type ePCR products gives two bands at sizes, 101 and 132 bp (shown by a star). C. ePCR analyses of leukemic cell lines demonstrate the presence of mutant gDNA in the MOLT-4 cell line (pointed by an arrow head) but not in other leukemic cell lines in this experiment. D. Direct sequence analysis of a gDNA ePCR product shows selective enrichment of the R46X mutation. E, F. Sequence chromatograms of the heterozygous DNA mutations detected in two T-ALL cell lines are shown.