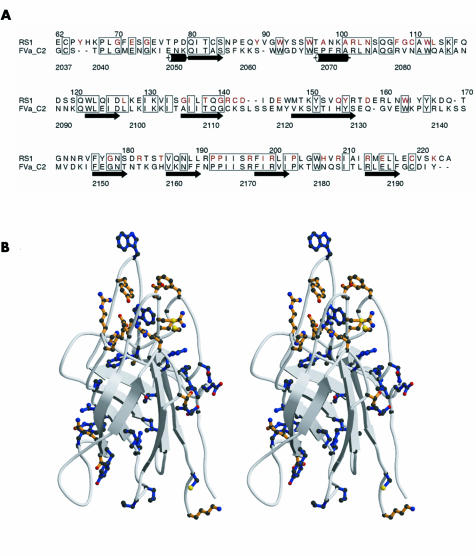
Figure 1 Homology based modelling of discoidin domain of retinoschisin. (A) Alignment of amino acid sequences of discoidin domains of retinoschisin (RS1) and factor Va (FVa_C2), prepared with Alscript. Boxes outline residues that are identical in the two sequences, retinoschisin residues mutated in disease are highlighted in red, and the secondary structure of the factor Va crystal structure19 indicated schematically. (B) Ribbon diagram (as stereo pair) of the homology based model of the retinoschisin discoidin domain showing the β barrel structure with long loops, prepared with Molscript (www.avatar.se/molscript) . The side chains shown indicate the positions of all mutations in retinoschisin patients to date. Sites of mutation that are conserved in the sequence alignment with factor Va are shown in blue, while mutation sites that are not conserved are shown in orange.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.