Fig. 1. Site-specific recombination mediated by Flp.
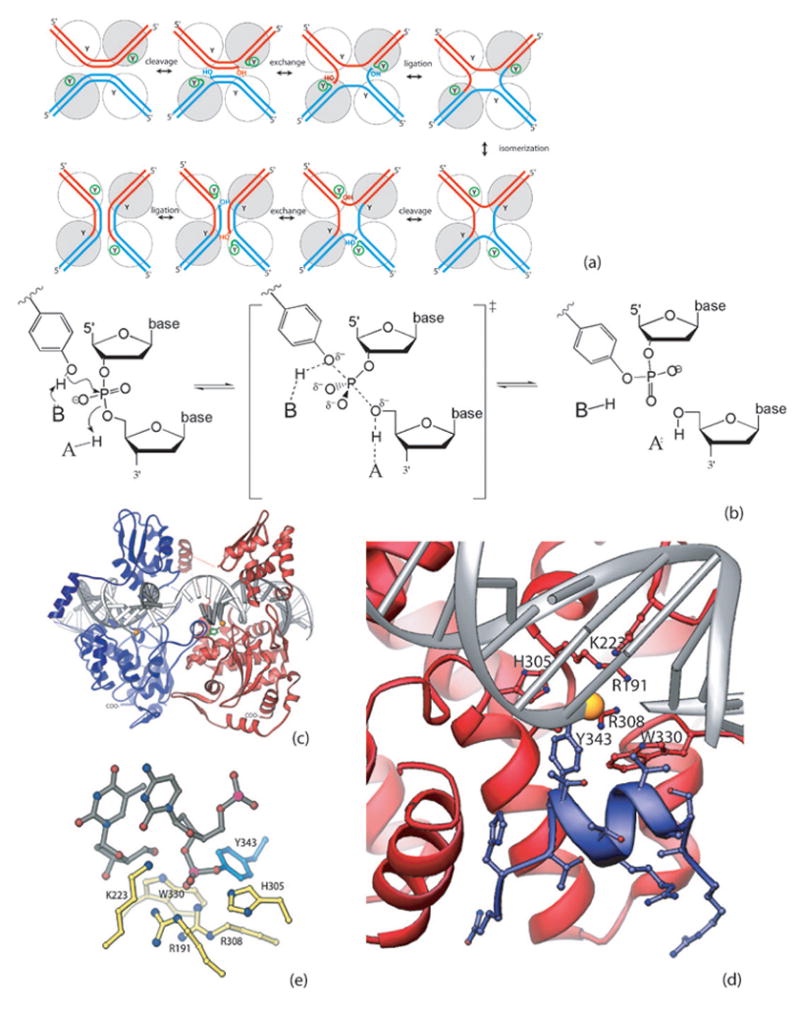
(a) Recombination pathway. Two DNA segments and four enzyme molecules (spheres, with Y representing the catalytic tyrosine) are required. Only two protomeres are active at each step, as indicated by the small circle around the Ys. The catalytic states of the individual protomers are switched in the isomerization step. The two DNA strands are sequentially cleaved, exchanged and religated. (b) General acid/base catalysis during the DNA cleavage and ligation reactions. In the DNA cleavage reaction, a general base presumably accepts a proton from the attacking tyrosyl hydroxyl group while a general acid donates a proton to the DNA’s 5’-bridging oxygen. This process is reversed in the ligation reaction. (c) Flp cleaves DNA in trans. The interface between two protomers is viewed from the center of the Holliday junction. Y343 of the blue protomer, shown in ball and stick mode, is donated to the red protomer and cleaves the DNA bound by that one. The scissile phosphate is shown as a gold sphere. (d) Flp Helix M (blue) packing into the active site of a neighboring Flp protomer (red). The segment shown corresponds to the peptide used in this study, and all side chains are shown. (e) Catalytic residues. The active state of the catalytic site, containing the covalent phosphotyrosine linkage, is shown. Six conserved residues, including Y343, surround the scissile phosphate. PDB code 1M6X
