Fig. 4.
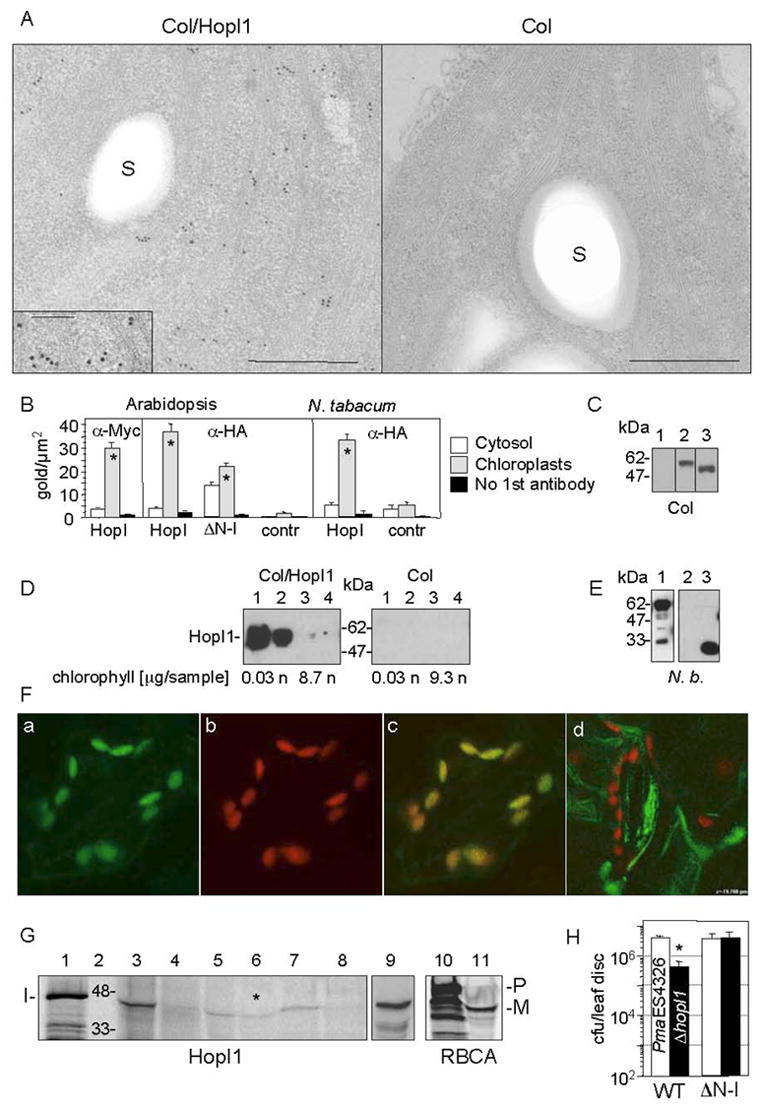
Subcellular chloroplast localization of HopI1 in transgenic HopI1 A. thaliana and transiently-transformed tobacco.
(A) Immunolocalization of constitutively expressed HopI1PmaES4326-HA-Myc-His in chloroplasts of 16 day old transgenic A. thaliana leaves (Col/HopI1=JJ30, left) and wild-type leaves (Col, right). HopI1 localized mainly to chloroplasts in transgenic plants, as visualized with anti-HA antibody. A fragment of chloroplast is shown; insert shows a higher magnification view (bar=500 nm; in the insert 200 nm); S, starch grain.
(B) Statistical analysis of the density of gold-label in HopI1-expressing plants and control plants, detected with anti-HA or anti-Myc antibodies. ΔN-HopI1PmaES4326-HA-Myc-His (JJ51) still localized to chloroplasts in transgenic A. thaliana. HopI1PmaES4326 (JJ30) localized to chloroplasts of transiently-transformed N. tabacum leaves (60 h after Agroinfection). * P<0.008, t-test. HopI, plants expressing HopI1-HA-Myc-His (JJ30). ΔN-I, plants expressing ΔN-HopI1-HA-Myc-His (JJ51). Contr, control plants (wild type (A. thaliana) or transformed with empty vector pCB302-3 (tobacco)).
(C) HopI1 alleles were expressed in transgenic A. thaliana, as shown by SDS-PAGE and Western blot analysis with anti-HA antibody: (1) A. thaliana Col, (2) JJ30=hopI1PmaES4326-HA-Myc-His, (3) JJ51=ΔN29hopI1PmaES4326-HA-Myc-His. Lines 1–3 are from one experiment.
(D) HopI1 localization was confirmed by chloroplast fractionation. HopI1-HA-Myc-His (Col/HopI1=JJ30) was detected in transgenic A. thaliana with anti-Myc antibody. Intact chloroplasts were isolated on a percoll gradient and further partitioned into stromal and membrane fractions. (1 and 2) Stroma (soluble chloroplast proteins) from chloroplasts not treated (1) or treated (2) with thermolysin. (3 and 4) Membranes (thylakoids and envelope) from chloroplasts not treated (3) or treated (4) with thermolysin. The presence of chlorophyll was used as a marker for the chloroplast membranes. HopI1 was present in chloroplast stroma: n, not analyzed; sample, amount of sample loaded on a gel.
(E) HopI1Pph1448A::GFP fusion is expressed in transiently transformed N. benthamiana, as shown with anti-GFP antibodies. Lines: (1) JJ163=hopI1Pph1448A-GFP (upper band), (2) pTA7001, (3) GFP (pAOV-GFP).
(F) Localization of HopI1Pph1448A-GFP in chloroplasts of transiently transformed N. benthamiana. GFP fluorescence (a) colocalized with chloroplast autofluorescence (b). (c) Merged image. (d) GFP was excluded from chloroplasts in N. benthamiana transiently transformed with the GFP control (pAOV-GFP).
(G) In vitro import of HopI1PmaES4326 into chloroplasts. Lanes: (1) Aliquot of HopI1 transcription/translation reaction. (2) Protein marker (kDa). (3 and 4) Supernatant (proteins outside chloroplasts) without (3) or with (4) thermolysin treatment. (5 and 6) Stroma (soluble chloroplast proteins) from chloroplasts not treated (5) or treated (6) with thermolysin after HopI1 import. Asterisk indicates that HopI1 was protected from proteolysis in stroma. (7 and 8) Membranes (thylakoids and envelope) from chloroplasts not treated (7) or treated (8) with thermolysin. (9) Mixture of (1) and (5) in 1:18 ratio. The proteins comigrate, indicating lack of transit peptide removal (10) Aliquot of preRBCA transcription/translation reaction. (11) Stroma from chloroplasts treated with thermolysin after RBCA import. A transit peptide was removed in mature RBCA protected from thermolysin. I, HopI1; P, preRBCA; M, mature RBCA. All lanes except (9) are from one gel. The gel exposure time for audioradiography was shorter for lanes 10 and 11.
(H) Col constitutively expressing HopI1PmaES4326 lacking its N-terminal 29 amino acids (ΔN-I, JJ51) rescued the virulence defect of the ΔhopI1 PmaES4326 strain. (*The growth of the ΔhopI1 strain was reduced compared to the PmaES4326 strain on wild-type plants, P<0.0001).
