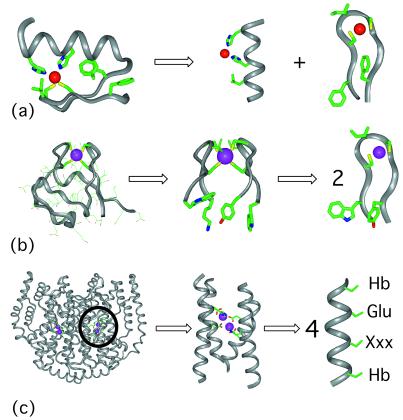
Figure 1.
Retrostructural analysis of metalloproteins. (a) A zinc finger from Zif268 residues 33 to 60 [PDB no. 1ZAA (61)]. The structure can be dissected in two separate secondary structure motifs including an idealized α-helix and β-hairpin. Shown are the two His and two Cys side chains critical for metal binding. Also shown are those hydrophobic resides that stabilize the fold as well as define the environment of the metal ion. (b Left) The full structure of rubredoxin [PDB no. 1BRF (84)], whose active site can be dissected in two symmetry-related idealized β-hairpins, residues 3–12, and 36–45 (Center). The β-hairpin from rubredoxin (Right) is remarkably similar to that found in Zif268 (a Right) in both its geometry and the placement of the aromatic and aliphatic side chains. (c Left) The full structure of Δ9 ACP desaturase [PDB no. 1AFR, (71)], which is a dimer of two identical catalytic subunits. In contrast to the complex nature of the overall structure, the architecture of the diiron-binding site can be described as a D2 symmetric four-helix bundle (Center). The idealized helix contains hydrophobic residues (Hb) that encapsulate and structurally stabilize the diiron site. Each helix contains a Glu side-chain ligand (shown). The position labeled “Xxx” is a His ligand in two of the helices. In the other two helices, this position helps define the environment and access surrounding the diiron center.