Figure 2.
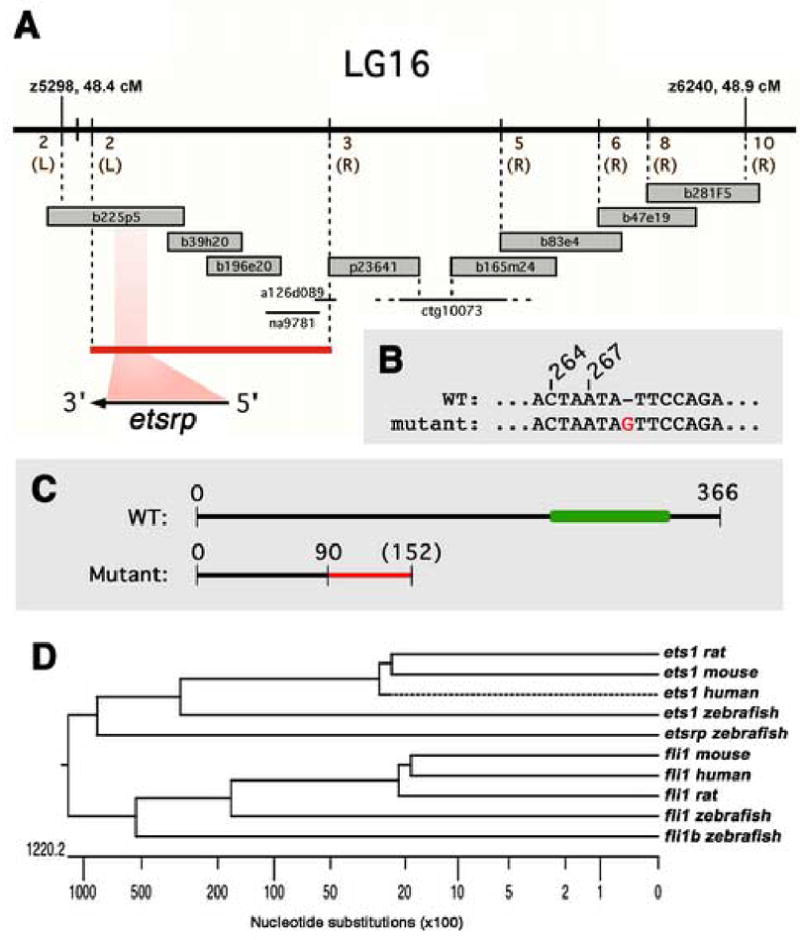
Positional cloning of etsrp, the defective gene in y11 mutants. (A) Genetic and physical map of the y11 interval. Genetic map is shown at top, with the number of recombinants in approximately 2000 meioses noted in brown (and whether recombinants are on the right or left sides, in parentheses). BAC and PAC clones are shown as large grey bars, genomic sequence contigs and traces are shown as black bars. The red bar shows the y11 critical interval containing the entire etsrp transcript. (B) In y11 mutants a G is inserted after nucleotide 269 in the etsrp coding sequence. (C) The y11 insertion mutation results in an altered reading frame after amino acid 90 of etsrp coding for an additional novel 62 amino acids (shown in red) before terminating at a stop codon. The y11 mutant protein lacks the highly conserved ETS DNA binding domain (shown in green in the wild type protein). See results and methods for additional details. (D) ETS family members expressed in the zebrafish vasculature and related mammalian orthologs. Dendogram of nucleotide simiilarity across the entire coding region of zebrafish ETS family members expressed in the zebrafish vasculature and the most closely related mammalian genes. The zebrafish ets1 and fli1 genes are more closely related to their mammalian counterparts than to other zebrafish ETS family members. The etsrp and fli1b genes are most closely related to ets1 and fli1 genes, respectively.
