Abstract
Proteins harboring a zona pellucida (ZP) domain are prominent components of vertebrate egg coats. Although less well characterized, the egg coat of the non-vertebrate marine gastropod abalone (Haliotis spp.) is also known to contain a ZP domain protein, raising the possibility of a common molecular basis of metazoan egg coat structures. Egg coat proteins from vertebrate as well as non-vertebrate taxa have been shown to evolve under positive selection. Studied most extensively in the abalone system, coevolution between adaptively diverging egg coat and sperm proteins may contribute to the rapid development of reproductive isolation. Thus, identifying the pattern of evolution among egg coat proteins is important in understanding the role these genes may play in the speciation process. The purpose of the present study is to characterize the constituent proteins of the egg coat [vitelline envelope (VE)] of abalone eggs and to provide preliminary evidence regarding how selection has acted on VE proteins during abalone evolution. A proteomic approach is used to match tandem mass spectra of peptides from purified VE proteins with abalone ovary EST sequences, identifying 9 of 10 ZP domain proteins as components of the VE. Maximum likelihood models of codon evolution suggest positive selection has acted among a subset of amino acids for 6 of these genes. This work provides further evidence of the prominence of ZP proteins as constituents of the egg coat, as well as the prominent role of positive selection in diversification of these reproductive proteins.
Keywords: adaptive evolution, egg coat proteins, gamete recognition
Metazoan eggs are surrounded by a fibrous coat referred to as the zona pellucida, as the vitelline or perivitelline envelope, or as the chorion. The constituent proteins of these structures, which we collectively refer to as egg coats, have been well characterized among vertebrate taxa, including species of mammals (1), teleost fish (2), amphibians (3), and birds (4). These studies show the principle constituents of vertebrate egg coats to be glycosylated proteins sharing a common structural motif of ≈260 aa known as the zona pellucida (ZP) domain. ZP domain proteins are found among diverse eukaryotic structures, facilitating protein polymerization through intramolecular disulfide bonds among conserved cysteine resides within the ZP domain (5). For example, the predominant model of mouse egg coat (zona pellucida) formation posits filaments of arrays of two ZP proteins (ZP2 and ZP3) crosslinked via a third (ZP1), resulting in a fibrous matrix completely enclosing the egg (1). Although orthologs of these mammalian proteins are known from egg coat structures across vertebrate lineages (6, 7), non-vertebrate taxa are less well studied and it is not yet clear whether ZP proteins are similarly prominent features of egg coats across metazoans. For example, all known proteins comprising Drosophila egg coat structures lack ZP domains (8). However, a large glycoprotein from the egg coat [vitelline envelope (VE)] of the marine gastropod abalone (Haliotis spp.) has been identified (9) and is known to contain a ZP domain (10), raising the possibility of a common molecular basis of animal egg coat structures.
As with reproductive proteins generally (11, 12), a remarkable feature of egg coat proteins is their diversity and rapid evolution as a result of positive Darwinian selection. A signature of positive selection has been found among ZP proteins from the egg coats of mammals (13) as well as abalone (10). Whereas the forces driving positive selection remain unclear (12), evidence is beginning to suggest adaptive divergence of egg coat proteins may be correlated with the evolution of interacting sperm proteins. For example, sites within ZP3 under selection among mammals (13) correspond to the putative sperm binding domain in mouse (14, 15). Similarly, the abalone sperm protein lysin binds to the ZP glycoprotein vitelline envelope receptor of lysin (VERL) (9) to facilitate sperm access to the egg membrane surface during fertilization (16), and both proteins show signs of adaptive divergence among abalone taxa (10, 17, 18). Coevolution between gamete recognition molecules is of significant interest because of their potential contribution to rapid reproductive isolation (19), which theoretical models show can occur even in sympatry if sexual conflict develops between components of male and female fitness (20). Thus identifying the selective forces acting on egg coat proteins is a preliminary step in establishing their potential contribution to the speciation process.
Whereas abalone represents one of the best systems for studying the causes and consequences of the evolution of interacting sperm and egg coat molecules, the constituent proteins of the abalone egg coat (VE) are less well described than for many vertebrate taxa. The purpose of the work described here is twofold: (i) to identify the major constituent proteins of the VE of abalone eggs; and (ii) to provide preliminary evidence of how selection has acted among these egg coat proteins. Addressing these issues sheds light on the questions of whether metazoan egg coat structures may share a common molecular basis (the ZP domain), and whether adaptive divergence is a common feature among abalone egg coat proteins. Toward these goals ≈5,000 randomly selected ESTs were sequenced from a pink abalone (Haliotis corrugata) ovary cDNA library. Peptide spectra from tandem mass spectrometry (MS/MS) of both pink and green abalone (Haliotis fulgens) VEs were matched to translated ESTs to identify abalone VE proteins. Orthologs were then sequenced from additional abalone taxa and maximum likelihood models of codon evolution used to test for evidence of positive selection among abalone VE proteins.
Results
Ovary-Expressed ZP Domain Genes.
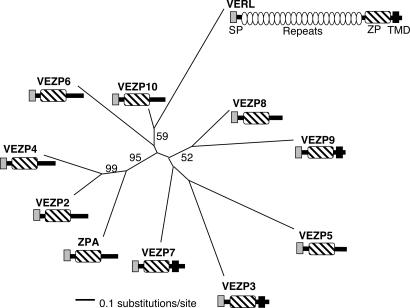
Of the ≈5,000 randomly selected clones sequenced, 50 (1%) assembled into 15 ESTs showing homology to ZP domain proteins. One of these was identical to the previously described pink VERL sequence (21). RACE successfully extended the coding sequence for 10 of the remaining 14 ESTs, providing putative full length transcripts (see below) for these genes (accession nos. DQ453710, DQ453714, DQ453718, DQ453722, DQ453726, DQ453730, DQ453734, DQ453738, DQ453742, DQ453746, and DQ453750). In contrast, repeated attempts to obtain additional sequence for the remaining 4 ESTs by using RACE were unsuccessful. Computational prediction of domains for the 10 recently identified ZP genes confirmed the presence of a canonical ZP domain with 10 conserved cysteine residues as well as a signal peptide among all genes, and identified transmembrane domains among three genes (Fig. 1). ZP proteins are generally secreted molecules (5), and the presence of signal peptides along with polyA tails among RACE cDNAs of these 10 genes is consistent with transcripts representing full length coding sequences. Interestingly, none of the ovary-expressed ZP genes found contained extensive repeat arrays as are present in VERL (Fig. 1), and instead were of similar length (321–508 aa) to mammalian egg coat ZP proteins (1).
Fig. 1.
ZP domain genes identified from a pink abalone (H. corrugata) ovary EST library. RACE was used to extend ESTs predicted to have homology to ZP domain proteins, identifying nine ovary-expressed genes in addition to VERL. Phylogenetic analysis of the ZP domain from these genes (≈768 nucleotides) employing likelihood was performed under the general time reversible model with four rate categories by using PAUP (38). Terminal branches in the tree are labeled with a proposed gene name specifying whether the protein is found within solubilized VE material (VERL, VEZP2 to VEZP9) or not (ZPA), and bootstrap support based on 100 bootstrap replicates is shown for nodes with ≥50% support. Protein domains previously characterized for VERL (48) or computationally predicted via SMART (www.embl-heidelberg.de) for recently identified ZP genes are shown, including signal peptides (SP), internal repeats, and ZP, as well as transmembrane domains (TMD).
Ovary-expressed ZP domain genes from pink abalone are well diverged from each other. Alignment of the ZP domains of the ten ZP genes for which we obtained putative full length transcripts and VERL show on average 66% of nonsynonymous (amino acid changing, dN) nucleotide positions as having substitutions among pairwise comparisons (range of 25–93%). Phylogenetic analysis based on this same alignment reveals a maximum likelihood gene tree with poorly supported topology in most cases (Fig. 1). Significantly, although the sister relationships among a clade of three genes in this tree are clear, there is no evidence of pairs of similarly diverged genes. This result suggests recent genome expansion (e.g., polyploidy) has not played a role in diversification of this gene family in abalone consistent with cytological evidence (22, 23).
Identification of Abalone VE Proteins.
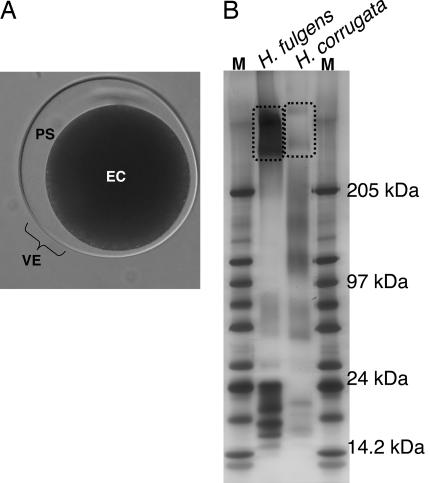
The VE of abalone eggs contains several proteins in addition to the large glycoprotein VERL (9). The majority of these are of significantly smaller size than VERL, migrating as a number of discrete peptides in the size range of 14.2 – 24 kDa (Fig. 2). Although the solubilized pink VE material also shows nondiscretely sized protein migrating in the size range of 97 – 250 kDa, this smear seems to be the result of degradation of higher molecular weight proteins such as VERL as (i) SDS/PAGE of freshly prepared solubilized VE material from pink abalone (9) seems qualitatively similar to the green abalone VE material (Fig. 2), and (ii) mass spectra of total solubilized pink as well as green VE material match a nearly identical set of sequences from the pink abalone ovary EST library (Table 1, and see below). MS/MS identified mass spectra for two or more peptides matched by SEQUEST to predicted amino acid sequences of only 18 (pink VEs) or 26 (green VEs) of the ESTs in the pink ovary library, with a false discovery rate ≤5%. Of these, 7 (pink VEs) or 9 (green VEs) correspond to the ovary-expressed ZP genes for which we obtained full coding sequence as well as VERL (Table 1). Because solubilized pink VE material shows evidence of degradation (Fig. 2), and the false discovery rate in these experiments is low (≤ 5%), this result is strong evidence that most (9 of 10) recently identified ZP domain genes are components of the VE in addition to VERL. We hereafter refer to these genes as VEZP2 to -10 (Table 1). The number of spectral counts for most of these proteins were qualitatively similar. However, spectral counts for VEZP9 from both pink and green VEs were markedly higher relative to other VE proteins including VERL, suggesting VEZP9 is the most abundant of these proteins. Interestingly, mass spectra for two or more peptides from both pink and green VEs also matched to the 4 ESTs in our ovary library showing evidence of homology to ZP domain proteins, but for which we were unable to confirm expression via RACE (data not shown). This result suggests several additional ZP proteins may ultimately be identified as components of the VE of abalone eggs.
Fig. 2.
Vitelline envelope proteins from abalone eggs. (A) Abalone eggs are surrounded by a raised VE separated from the egg (EC, egg cytoplasm) by a perivitelline space (PS). (B) Five micrograms of total solubilized protein from purified VEs of both green (H. fulgens) and pink (H. corrugata) abalone were run on 2.5–15% SDS/PAGE gels and silver stained. The high molecular mass bands (boxed) identified as VERL (9) were confirmed by MS/MS of gel slices. Approximate molecular masses of size standards (M) are shown to the right. Aliquots of the same total VE proteins from both species were used for peptide identification in MS/MS experiments.
Table 1.
Constituent ZP proteins of the VE of abalone eggs
| Gene | VE Mass Spectra* |
|
|---|---|---|
| H. corrugata | H. fulgens | |
| VERL | 1 (1) | 4 (5) |
| VEZP2 | 1 (1) | 2 (2) |
| VEZP3 | 0 (0) | 2 (5) |
| VEZP4 | 0 (0) | 6 (27) |
| VEZP5 | 2 (3) | 2 (10) |
| VEZP6 | 4 (57) | 2 (11) |
| VEZP7 | 1 (2) | 3 (12) |
| VEZP8 | 3 (50) | 4 (2) |
| VEZP9 | 12 (122) | 6 (177) |
| VEZP10 | 1 (1) | 4 (12) |
| ZPA | 0 (0) | 0 (0) |
Solubilized VE material (Fig. 1) from eggs of pink (H. corrugata) and green (H. fulgens) abalone were subjected to MS/MS, and mass spectra were matched by SEQUEST to the pink abalone EST library. The number of unique peptides and total number of spectral counts for unique peptides (in parentheses) matching to each of 11 ovary-expressed ZP domain genes are shown. Results from mass spectrometry confirm the presence of VERL, known to be a major component of the VE (49) as well as identifying 9 of 10 recently identified ZP domain proteins (VEZP2 to VEZP10) as constituents of the VE. Based on the relative abundance of spectral counts, which can be used as a proxy for the relative abundance of peptides (44), VEZP9 appears to be the most abundant of these proteins. ZP domain proteins from the EST library not matched to VE peptides are given an arbitrary alphabetical designation (ZPA).
*5% false discovery rate.
Adaptive Divergence Among Abalone ZP Orthologs.
Orthologs of all 10 recently identified pink abalone ZP genes as well as the C terminus portion of VERL were cloned from red (H. rufescens), green, and Japanese ezo (H. discus hannai) abalone employing our modified RACE approach (accessions DQ453711 to DQ453713, DQ453715 to DQ453717, DQ453719 to DQ453721, DQ453723 to DQ453725, DQ453727 to DQ453729, DQ453731 to DQ453733, DQ453735 to DQ453737, DQ453739 to DQ453741, DQ453743 to DQ453745, and DQ453747 to DQ453750). Although the relationships among gene clades are generally weakly supported in phylogenetic analyses of all 44 sequences (data not shown), similar to our earlier results (Fig. 1), in all but one case each ZP gene clade is strongly supported consistent with orthology (≥99% bootstrap support). The exception is a monophyletic clade containing the four putative VEZP10 orthologs that is less strongly supported, but for which bootstrap support is still high (80%). Regardless, in all cases substitutions among orthologs at nonsynonymous and synonymous (dS) nucleotide positions are similar and limited (averaging 3% and 7% among pairwise comparisons, respectively), consistent with the recent diversification of these taxa (<18 million years ago) (24).
Despite the limited power of the codon substitution models when used to detect adaptive divergence among such closely related taxa (25), we found evidence of positive selection acting on 6 of 10 recently identified ZP genes. A signature of positive selection on coding regions is evident from the proportionally higher dN relative to dS, resulting in a dN/dS ratio (ω) >1. Model M0 allowing for a single ω among all sites suggests positive selection has driven the divergence of two ZP genes (VEZP5, VEZP9; Table 2). Likelihood ratio tests comparing more powerful selection models (M2a, M8) with a corresponding nested neutral model (M1a, M7 or M8a, respectively) that allow for variable ω ratios among sites consistently show the selection model resulting in a significantly better fit to the data for 6 of 10 ZP genes including VEZP3 to -6 and VEZP9 to -10. Although Table 2 presents results only for models M8a and M8, for which model comparisons by using likelihood ratio tests are the most conservative and robust (26), results from other neutral and selection model comparisons are qualitatively similar. Model M8 shows a small proportion of sites under selection (ω>1) for each of these genes (1–11%), again consistent with results from other selection models. Significantly, simulations show the LRT applied to these model tests is robust and does not lead to an elevated type 1 error rate even for the small sample size used here (25).
Table 2.
Tests of positive selection among abalone ZP genes
| Gene | One ratio model (M0) | Neutral model (M8a) | Selection model (M8) | 2 Δ lnL (M8a-vs-M8) |
|---|---|---|---|---|
| VERL | ω0 = 0.5 | ω0 = 1.00, pO = 0.55 | ω0 = 2.34, pO = 0.14 | 0.70 |
| (C terminus only) | ||||
| VEZP2 | ω0 = 0.20 | ω0 = 1.00, pO = 0.10 | ω0 = 1.00, pO = 0.10 | 0 |
| VEZP3 | ω0 = 0.60 | ω0 = 1.00, pO = 0.48 | ω0 = 3.7, pO = 0.08 | 6.24* |
| VEZP4 | ω0 = 0.40 | ω0 = 1.00, pO = 0.40 | ω0 = 3.5, pO = 0.11 | 5.86* |
| VEZP5 | ω0 = 1.06 | ω0 = 1.00, pO = 0.56 | ω0 = 10.16, pO = 0.07 | 28.64** |
| VEZP6 | ω0 = 0.17 | ω0 = 1.00, pO = 0.51 | ω0 = 21.34, pO = 0.01 | 6.20* |
| VEZP7 | ω0 = 0.57 | ω0 = 1.00, pO = 0.46 | ω0 = 1.00, pO = 0.46 | 0 |
| VEZP8 | ω0 = 0.45 | ω0 = 1.00, pO = 0.00 | ω0 = 1.00, pO = 0.00 | 0 |
| VEZP9 | ω0 = 1.11 | ω0 = 1.00, pO = 0.80 | ω0 = 10.06, pO = 0.04 | 9.22* |
| VEZP10 | ω0 = 0.11 | ω0 = 1.00, pO = 0.10 | ω0 = 10.74, pO = 0.01 | 7.96* |
| ZPA | ω0 = 0.17 | ω0 = 1.00, pO = 0.16 | ω0 = 1.74, pO = 0.11 | 1.12 |
Codon substitution models (46, 47) were used to analyze sequences from four abalone taxa (H. corrugata, H. rufescens, H. fulgens, and H. discus hannai). Sites models allowing for a single ω across sites (M0), as well as several neutral models (M1a, M7, and M8a) or selection models (M2a and M8) allowing for variation in ω among sites, were fit to the data using PAML (40). Despite low power of M0 to detect positive selection, two VE proteins (VEZP5 and VEZP9) show evidence of ω > 1 consistent with adaptive divergence among species. More powerful tests employing likelihood ratio tests comparing neutral and selection models (e.g., M8a-vs-M8) identify four additional VE proteins under positive selection (VEZP3, VEZP4, VEZP6, and VEZP10). Estimates of ω, the proportion of codons in this positively selected site class (ω 0 and p0, respectively), and the likelihood ratio test statistic (LRT, 2 Δ lnL) are given. ∗, significant at P ≤ 0.05; ∗∗, significant at P ≤ 0.005.
Discussion
The work presented here has two major findings. First, like many vertebrate taxa, ZP domain proteins are major constituents of the egg coat of non-vertebrate marine gastropod abalone. Second, a signature of positive selection common among reproductive proteins generally is a prominent force driving the adaptive diversification of the VE proteins identified among abalone species. These results are discussed in detail below, including the significance of our findings toward a broader understanding of reproductive protein diversity.
Abalone and Vertebrate Egg Coats Have a Common Molecular Basis.
The eggs of metazoan animals are surrounded by a fibrous coat variously referred to as the zona pellucida, vitelline or perivitelline envelope, or chorion. The composition of these structures, which we collectively refer to as egg coats, have been studied most extensively among vertebrate taxa. Initial biochemical characterization in mouse (27) identified three glycosylated proteins as the principle components of the egg coat. These genes were shown to share a motif of ≈260 aa coined the zona pellucida (ZP) domain after the structure from which they were isolated. More recent biochemical and proteomics studies of egg coats from other mammals (including humans) have identified orthologs of mouse ZP1-3, as well as a fourth ZP protein apparently lost in the mouse lineage (28). Phylogenetic analyses of these mammalian genes along with homologues from birds, fish, and amphibians (4, 6) suggests five to six ancestral ZP genes were present before diversification of the vertebrates, with subsequent lineage-specific loss (e.g., mouse) or gain (e.g., teleost fish). Taken together, the consensus beginning to emerge from these studies is that vertebrate egg coat structures are comprised of a core set of just a few orthologous ZP proteins (three to four in higher mammals).
We find that ZP domain proteins are also prominent components of the abalone egg coat. The MS/MS studies of abalone VEs identified multiple peptides for 7 (pink abalone) or 9 (green abalone) of 10 fully sequenced ZP domain genes found in the ovary EST library as well as VERL (Table 1). Peptides were also identified from VEs and matched to each of four distinct but uncharacterized ESTs from the library showing homology to ZP domain proteins via BlastX. Thus the abalone egg coat seems to contain at least 10 ZP domain proteins (VERL and VEZP9 to -10), with the possibility of several more as yet uncharacterized ZP proteins also present. All of these genes are well diverged with respect to each other having on average 66% of nonsynonymous nucleotide positions with substitutions within the ZP domain (similar to the divergence between mouse ZP1 and ZP2), and there is no evidence from the gene phylogeny (Fig. 1) or cytological evidence (22, 23), suggesting recent polyploidy is responsible for the diversity of these genes. This finding makes the abalone VE one of the most complex egg coat structures described to date in terms of the number of ZP domain proteins present.
There are two distinct explanations for the shared prominence of ZP domain proteins among egg coat structures from abalone and vertebrates. First, metazoan egg coat structures might share a common evolutionary origin. Although intriguing, we currently find no support for this hypothesis as all of the abalone ZP domain proteins characterized here form a well supported monophyletic clade along with VERL distinct from vertebrate egg coat proteins (data not shown). In addition, all known proteins present within Drosophila egg coat structures lack a ZP domain (8). Second, the ZP domain may have been independently recruited more than once during metazoan evolution for its structural role in the synthesis of the egg coat matrix. ZP domain proteins are found as components of diverse eukaryotic structures, and seem to play a key structural role in the formation of filaments and membranes by facilitating protein polymerization through intramolecular disulfide bonds (5). Under this scenario, the prevalence of ZP domain proteins in the abalone egg coat represents another example of the utility of this structural motif during metazoan evolution. Distinguishing between these hypotheses will require further characterization of egg coat molecules among representative taxa from other non-vertebrate lineages. Regardless, a common molecular basis suggests vertebrates and non-vertebrates may share general features of fertilization involving molecular interactions with the egg coat.
Positive Selection Drives the Evolution of Most Abalone Egg Coat Proteins.
A common theme among reproductive proteins is their diversity and rapid divergence as a result of positive selection (11). This pattern has been demonstrated previously for proteins functioning at qualitatively distinct stages of reproduction including fertilization (12), with examples from several mammalian egg coat proteins as well as abalone VERL. Comparison of >2,800 human-mouse orthologs identified ZP1, ZP2, and ZP3 as among the 10% most divergent genes (29). A subset of sites among mammalian orthologs of ZP2 and ZP3 under selection have been identified (13), and preliminary analyses of primate ZP1 orthologs also show evidence of positive selection (W.J.S., unpublished data). These studies demonstrate positive selection has been a potent evolutionary force contributing to the rapid divergence of egg coat proteins among mammals. Similarly, although a majority of the repeat array of VERL evolves through a process of gene conversion (21) and there is no evidence of positive selection acting within the C terminus of the protein in our study (Table 2) or previously (30), a signature of positive selection has been detected previously within the two N terminus VERL repeats (10).
The results of the current work examining divergence of the recently identified VE ZP genes among abalone taxa complement previous studies of VERL (10), providing one of the most comprehensive descriptions of the selective pressures acting on the constituent proteins of an egg coat structure. Of 9 recently identified VE proteins (Table 1), we found evidence of positive selection acting at a subset of sites for 6 genes (VEZP3 to -6, VEZP9 to -10; Table 2). Although statistical power of the codon substitution models we employ to detect positive selection is low for the small sample sizes used here, the rate at which false cases of positive selection are detected should not be elevated (25). Thus we have strong evidence that positive selection has contributed to the divergence of the majority (70%) of the known protein components of the VE among abalone taxa (all containing a ZP domain).
The prevalence of positive selection among abalone VE proteins is remarkable, and raises the question of how selective forces may be acting to drive adaptive diversification so broadly among constituent egg coat proteins. Although the specific selective forces remain unclear, several causes have been proposed to act singly or in combination which may contribute to this pattern (11, 12). First, microbial attack of gametes can impose constant selective pressure among gamete surface proteins to change to elude pathogens (19). Selection pressures from pathogens are likely to occur among both free-spawning taxa encountering pathogens in the environment such as abalone, and in internal fertilizers because of sexually transmitted pathogens. Second, sperm competition (31) can result in selection on sperm proteins because sperm compete individually for first access to the egg. When sperm are abundant, this race can result in sexual conflict (32) which reflects the competing interests between male and female reproduction. For example, if multiple sperm fuse with an egg (polyspermy), development will arrest resulting in strong selection on eggs to slow sperm access or blocks to polyspermy (33). The physical barrier to sperm entry presented by the structural assembly of egg coat ZP proteins may represent a broad set of targets for such a mechanism.
Additional explanations for positive selection on gamete proteins have been proposed, although it is unclear whether they may represent a general mechanism driving egg coat protein evolution. Sexual selection at a cellular level can drive coevolution between egg and sperm proteins. For example, sea urchin eggs (Echinometra spp.) show a clear preference for sperm carrying their own allele of the sperm protein bindin (34). And although selection for reproductive isolation to avoid low fitness matings remains controversial (35), reinforcement has similarly been proposed as a potential driving force of reproductive protein evolution. Because ZP egg coat proteins are thought to contribute to heterospecific barriers to fertilization (refs. 1 and 10; and see following paragraph), reinforcement is a possible source of positive selection.
Regardless of the cause, a significant consequence of the diversification of egg coat proteins under positive selection is reproductive isolation because of incompatibilities with interacting sperm proteins during fertilization. For example, although the mechanism of sperm recognition is in debate (1), a portion of sites within the mammalian egg coat protein ZP3 predicted to be under positive selection (13) correspond to the putative sperm binding domain of ZP3 from mouse (14, 15). Because ZP3 is thought to be responsible for the heterospecific barrier to fertilization presented by the mammalian egg coat through its sperm receptor activity (1), this result suggests adaptive divergence of ZP3 might contribute to reproductive isolation. The strongest evidence for the role of egg coat proteins in reproductive isolation come from abalone, where the sperm protein lysin facilitates sperm access to the egg membrane surface by dissolving a hole through the elevated egg VE (16) through binding to VERL (9). Lysin domains under positive selection are responsible for species-specific dissolution of the vitelline envelope (36), and are believed to coevolve with a subset of repetitive VERL elements similarly found to be under positive selection among abalone species (10). Adaptive divergence of lysin and VERL are consistent with a “coevolutionary chase” between male and female reproductive proteins, which theoretical models (20) show can result in reproductive isolation among genotypes in sympatry when sexual conflicts develop between male and female components of fitness. Taken together with evidence from ZP3, this observation suggests positive selection on egg coat proteins may represent a primary and critical step contributing to the speciation process.
Function of Abalone VEZP Proteins.
The specific function of the nine recently identified VE ZP proteins within the abalone egg coat have not yet been established. However, the presence of a ZP domain within all of these genes (Fig. 1) suggests they play some role in the structural assembly of the VE, perhaps facilitating protein polymerization to form a matrix of ZP proteins similar to that proposed for the mouse model (1). The abalone VE is known to be a complex structure, with at least three layers composed of fibers with distinct dimensions and conformations (37). It will be interesting in future studies to identify the distribution and relative proximity of VEZP proteins throughout these layers. Although it seems unlikely that VEZP2 to -9 interact directly with sperm lysin because of the lack of homology with VERL repeats (Fig. 1) where lysin is believed to bind (11), it will also be interesting to establish whether these proteins function in binding to sperm membrane proteins or triggering the acrosome reaction, as these processes are known to occur at the VE surface (37). If interactions with sperm proteins are found, the prevalence of positive selection among VE ZP proteins suggests several loci in addition to lysin-VERL are promising candidates for further study of the molecular basis of reproductive isolation among abalone taxa.
Conclusions
We find that ZP domain proteins are prominent components of the egg coat (VE) of non-vertebrate marine gastropod abalone, which is comprised of at least 9 ZP domain proteins in addition to VERL. A signature of positive selection is found within 6 of the recently identified VE ZP genes, consistent with the adaptive diversification of most (7 of 10) known VE proteins among abalone species. A common molecular basis of egg coats employing the ZP structural domain suggests vertebrate and non-vertebrate animals may also share general features of fertilization that involve molecular interactions with egg coat structures. This finding suggests marine non-vertebrates such as abalone are valuable model systems for studying the evolutionary forces driving the common pattern of positive selection on egg coat proteins, and how divergence of these molecules may contribute to reproductive isolation.
Materials and Methods
Abalone Ovary ESTs.
An abalone ovary library of ESTs was generated by randomly sequencing ovary cDNAs. Total RNA was prepared from pink abalone (H. corrugata) ovary tissue by using the guanidinium/cesium chloride centrifugation method (50), from which mRNA was isolated by using the Oligotex mRNA kit (Qiagen, Valencia, CA). Approximately 1 μg of this mRNA was used in the Gateway cDNA synthesis and cloning kit (Invitrogen, Carlsbad, CA) employing the pSPORT1 vector. Approximately 5,000 clones were randomly selected for plasmid DNA purification by using the PerfectPrep kit (Eppendorf, Hamburg, Germany) and were unidirectionally sequenced by using the M13 forward primer and standard fluorescent sequencing methods (Applied Biosystems, Foster City, CA). Traces were processed and assembled by using default parameters in PHREDPHRAP (www.phrap.org). Assembled sequences (ESTs) were used to search the National Center for Biotechnology Information (NCBI) nonredundant protein database by using the NCBI BLAST client server Blastcl3 (www.ncbi.nih.gov) for BlastX E values <10−5.
To determine whether all ESTs showing homology to ZP domain proteins correspond to distinct ZP genes, full coding sequences were obtained by using a RACE approach. Both 5′- and 3′-RACE cDNAs were synthesized from 1 μg of total pink ovary RNA by using the SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA). 5′- and 3′-RACE PCR primers were designed for each EST with hits to ZP domain proteins and used in SMART PCR following the manufacturers instructions (Clontech). RACE PCR products were cloned by using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and at least four clones from each PCR were sequenced as above. Sequences from RACE clones and ESTs were assembled as described previously for abalone ovary ESTs, and the presence of the ZP as well as other domains was confirmed via SMART (www.embl-heidelberg.de). Nucleotide alignments of the ZP domains from these genes were carried out initially based on the translated nucleotide (protein) sequence by using the ClustalX algorithm implemented in BioEdit (T. Hall, North Carolina State University, Raleigh), followed by visual alignment. Areas with ambiguous alignments were excluded, and the nucleotide alignment used in phylogenetic analyses employing likelihood criterion implemented in PAUP (38). Likelihood analyses used the general time reversible model with four rate categories, estimating the γ shape parameter and the proportion of invariable sites (GTR + 1 + γ). Heuristic search criteria included tree bisection-reconnection (TBR) branch swapping with 10 random addition replicates. Support for nodes was estimated by 100 bootstrap replicates by using the maximum likelihood estimates of substitution parameters and heuristic search criterion. Pairwise nonsynonymous (dN) and synonymous (dS) nucleotide substitutions for this alignment were calculated employing maximum likelihood in the PAML computer package (39).
Identification of VE Proteins.
Constituent proteins of the abalone egg VE were identified by using tandem mass spectrometry (MS/MS). VE proteins from pink as well as green abalone (H. fulgens) prepared from whole eggs following the protocol of (16) were provided as a gift from V. Vacquier (The Scripps Institute of Oceonography, La Jolla, CA). VEs were solubilized following the protocol of ref. 9, aliquots of solubilized protein were reserved for SDS/PAGE, and the remainder of VE proteins were reserved for MS/MS. For SDS/PAGE, 5 μg of total pink and green VE proteins were loaded, run, and silver stained on 2.5–15% acrylamide gels as in ref. 9.
For MS/MS, the protein was denatured with 0.1% Rapigest (Waters Corporation, Milford, MA) in 50 mM ammonium bicarbonate buffer, reduced with 5 mM DTT, and alkylated with 15 mM iodoacetamide. The protein was then digested with trypsin at a 1:100 enzyme:substrate ratio and incubated with mixing at 37°C for 4 h. At the end of the 4 h, the digest was quenched, and the Rapigest was hydrolyzed with the addition of 200 mM HCl. The hydrolyzed Rapigest was pelleted by centrifugation. The protein digests were then loaded directly onto a fused silica capillary column (100 μM i.d.) packed with 7 cm of 5 μm Luna C18 material (Phenomenex, Ventura, CA) at the tip, followed by an additional 3 cm of 5 μM Luna C18 material as in ref. 40. After loading the peptide digests, the column was placed inline with an Agilent 1100 HPLC/Autosampler and analyzed by using a six-step multidimensional separation similar to ref. 41. The HPLC was run at 150 μl/min and split to ≈200 nl/min immediately upstream of the capillary column. The peptides were displaced from the strong cation exchange (SCX) resin onto the reversed phase material by using six separate salt fractions consisting of a 50-μl injection of ammonium acetate from the autosampler (0 mM, 100 mM, 200 mM, 500 mM, 800 mM, and 5,000 mM). Each salt injections was followed by a 2-h water:acetonitrile gradient. As peptides eluted from the microcapillary column, they were electrosprayed directly into an LTQ linear ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA) with the application of a distal 2-kV spray voltage. A cycle of one full-scan mass spectrum (400–1400 m/z) followed by five data-dependent MS/MS spectra at a 25% normalized collision energy repeated continuously throughout each step of the multidimensional separation. The application of all mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system.
The acquired tandem mass spectra were searched against a database containing the EST 6 reading frame translations, all known Haliotis protein sequences from NCBI, proteins of common contaminants (e.g., trypsin and keratin), and a shuffled decoy database by using a parallelized implementation of SEQUEST-NORM (41) with no enzyme specificity selected in the parameter file. The program DTASelect (42) was used to filter the peptide identifications and assemble the peptides and proteins. DTASelect filters were adjusted to produce protein identifications with a false discovery rate of ≤5%. The relative abundance of VE peptides was inferred from the number of spectral counts, which has been shown to approximate the relative abundance of peptides identified in MS/MS studies of complex mixtures of peptides (43).
Tests of Positive Selection Among ZP Genes.
To test for evidence of positive selection among ovary-expressed ZP genes, orthologs were cloned from three additional abalone species. Ovary total RNA was prepared from green, red (Haliotis rufescens), and Japanese ezo (Haliotis discus hannai) abalone and used to construct 5′- and 3′-RACE cDNAs as described previously for red abalone. For all ZP genes identified except VERL, degenerate 3′-RACE primers were designed from complete pink ZP gene sequences (primers available from authors upon request), used in 3′-RACE from cDNA of each species following the manufacturer's guidelines (Clontech, Palo Alto, CA), and RACE PCRs were TOPO cloned and sequenced as described previously for red abalone. Gene specific (nondegenerate) primers were then designed from species-specific 3′-RACE sequences and used in 5′-RACE. For VERL, the entire nonrepetitive C terminus sequence (10) was obtained through 3′-RACE alone by using a degenerate primer (available from authors upon request) or was publicly available (H. rufescens; accession no. AF453553). To confirm orthology, the ZP domains from all 44 sequences were aligned and analyzed simultaneously by using maximum likelihood criteria to determine phylogenetic relationships among genes, and pairwise dN and dS among each of the 11 sets of orthologs were calculated for the entire coding region by using maximum likelihood as described previously for pink abalone ZP genes.
We looked for a signature of positive selection among abalone ZP genes by comparing dN with dS for all 11 sets of orthologs. The ratio (dN/dS, or ω) can be used as an index of selection where ω < 1 is consistent with purifying selection, ω = 1 indicates neutral evolution, and ω > 1 is consistent with positive selection (i.e., adaptive diversification of orthologs). Codon substitution models employing maximum likelihood (44) including sites models (45, 46) that allow for variable ω ratios among codons were used to individually analyze all 11 sets of abalone orthologs after removing signal peptides and manually aligning the remaining full coding sequences. In addition to a model allowing for a single ω ratio among all codons (M0), several neutral (M1a, M7, M8a) and selection (M2a, M8) models were fit to the data by using the computer program PAML (39). An unrooted phylogeny placing pink and Japanese ezo abalone as sister (47) was used for all models, and those employing a β distribution (M7, M8a, and M8) included 10 rate categories. Likelihood ratio tests of nested neutral and selection models were compared with χ2 distributions with one (M8a-vs.-M8) or two (M1a-vs.-M2a, M7-vs.-M8) degrees of freedom to establish statistical support for the added parameters of the respective selection model.
Acknowledgments
We thank V. Vacquier for providing abalone VE samples. This work was supported by National Institutes of Health (NIH) Grant HD42563 and National Science Foundation Grants DEB-0410112 and DEB-0213171 (to W.J.S.), by NIH Grant P41 RR011823 (to M.J.M.), and by NIH Institutional Training Grant T32 HG00035 (to J.E.A.).
Abbreviations
- ZP
zona pellucida
- VE
vitelline envelope
- MS/MS
tandem mass spectrometry
- VERL
vitelline envelope receptor of lysin.
Footnotes
References
- 1.Wassarman PM, Jovine L, Qi HY, Williams Z, Darie C, Litscher ES. Mol Cell Endocrinol. 2005;234:95–103. doi: 10.1016/j.mce.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Darie CC, Biniossek ML, Jovine L, Litscher ES, Wassarman PM. Biochemistry. 2004;43:7459–7478. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- 3.Barisone GA, Albertali IE, Sánchez M, Cabada MO. Reprod Biol Endocrinol. 2003;1:18. doi: 10.1186/1477-7827-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith J, Paton IR, Hughes DC, Burt DW. Mol Reprod Dev. 2005;70:133–145. doi: 10.1002/mrd.20197. [DOI] [PubMed] [Google Scholar]
- 5.Jovine L, Darie CC, Litscher ES, Wassarman PM. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- 6.Spargo SC, Hope RM. Biol Reprod. 2003;68:358–362. doi: 10.1095/biolreprod.102.008086. [DOI] [PubMed] [Google Scholar]
- 7.Conner SJ, Lefievre L, Hughes DC, Barratt CL. Hum Reprod. 2005;20:1148–1152. doi: 10.1093/humrep/deh835. [DOI] [PubMed] [Google Scholar]
- 8.Waring GL. Int Rev Cytol 198. 2000:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- 9.Swanson WJ, Vacquier VD. Proc Natl Acad Sci USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galindo BE, Vacquier VD, Swanson WJ. Proc Natl Acad Sci USA. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson WJ, Vacquier VD. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 12.Clark NL, Aagaard JE, Swanson WJ. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- 13.Swanson WJ, Yang Z, Wolfner MF, Aquadro CF. Proc Natl Acad Sci USA. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Litscher ES, Wassarman PM. Proc Natl Acad Sci USA. 1998;95:6193–6197. doi: 10.1073/pnas.95.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinloch RA, Sakai Y, Wassarman PM. Proc Natl Acad Sci USA. 1995;92:263–267. doi: 10.1073/pnas.92.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis CA, Talbot CF, Vacquier VD. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- 17.Lee YH, Ota T, Vacquier VD. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Swanson WJ, Vacquier VD. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
- 19.Vacquier VD. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- 20.Gavrilets S, Waxman D. Proc Natl Acad Sci USA. 2002;99:10533–10538. doi: 10.1073/pnas.152011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swanson WJ, Vacquier VD. Science. 1998;281:710–712. doi: 10.1126/science.281.5377.710. [DOI] [PubMed] [Google Scholar]
- 22.Thiriot-Quiévreux C. J Molluscan Stud. 2003;69:187–202. [Google Scholar]
- 23.Gallardo-Escarate C, Alvarez-Borrego J, Del Rio Portilla M, Kober V. J Shellfish Res. 2004;23:205–211. [Google Scholar]
- 24.Metz EC, Robles-Sikisaka R, Vacquier VD. Proc Natl Acad Sci USA. 1998;95:10676–10681. doi: 10.1073/pnas.95.18.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anisimova M, Bielawski JP, Yang Z. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- 26.Wong WS, Yang Z, Goldman N, Nielsen R. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bleil JD, Wassarman PM. Dev Biol. 1980;76:185–202. doi: 10.1016/0012-1606(80)90371-1. [DOI] [PubMed] [Google Scholar]
- 28.Lefievre L, Conner SJ, Salpekar A, Olufowobi O, Ashton P, Pavlovic B, Lenton W, Afnan M, Brewis IA, Monk M, et al. Hum Reprod. 2004;19:1580–1586. doi: 10.1093/humrep/deh301. [DOI] [PubMed] [Google Scholar]
- 29.Makalowski W, Boguski MS. Proc Natl Acad Sci USA. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson WJ, Aquadro CF, Vacquier VD. Mol Biol Evol. 2001;18:376–383. doi: 10.1093/oxfordjournals.molbev.a003813. [DOI] [PubMed] [Google Scholar]
- 31.Clark AG, Begun DJ, Prout T. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- 32.Rice WR, Holland B. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 33.Gould-Somero M, Jaffe LA. In: Cell Fusion and Transformation. Beers RF, Bassett EG, editors. New York: Raven Press; 1984. pp. 27–38. [Google Scholar]
- 34.Palumbi SR. Proc Natl Acad Sci USA. 1999;96:12632–12637. doi: 10.1073/pnas.96.22.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noor MA. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- 36.Lyon JD, Vacquier VD. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- 37.Mozingo NM, Vacquier VD, Chandler DE. Mol Reprod Dev. 1995;41:493–502. doi: 10.1002/mrd.1080410412. [DOI] [PubMed] [Google Scholar]
- 38.Swofford DL. Sunderland, MA.: Sinauer Associates; 2002. PAUP: Phylogenetic Analysis Using Parsimony (And Other Methods) Version 4.0b10. [Google Scholar]
- 39.Yang Z. CABIOS. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 40.Wu CC, MacCoss MJ. Curr Opin Mol Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- 41.MacCoss MJ, Wu CC, Yates JR., 3rd Anal Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 42.Tabb DL, McDonald WH, Yates JR., 3rd J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 44.Goldman N, Yang Z. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen R, Yang Z. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Nielsen R, Goldman N, Pedersen AM. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coleman AW, Vacquier VD. J Mol Evol. 2002;54:246–257. doi: 10.1007/s00239-001-0006-0. [DOI] [PubMed] [Google Scholar]
- 48.Galindo BE, Moy GW, Swanson WJ, Vacquier VD. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. [DOI] [PubMed] [Google Scholar]
- 49.Vacquier VD, Swanson WJ, Lee YH. J Mol Evol. 1997;44:S15–S22. doi: 10.1007/pl00000049. [DOI] [PubMed] [Google Scholar]
- 50.MacDonald RJ, Swift GH, Przybyla AE, Chirgwin JM. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]