Abstract
Background
For breast and prostate cancer, a gene expression signature of the tumour is associated with the development of distant metastases. Regarding head and neck squamous cell carcinoma (HNSCC), the only known risk factor is the presence of ⩾3 tumour‐positive lymph nodes.
Aim
To evaluate whether a HNSCC gene expression signature can discriminate between the patients with and without distant metastases.
Methods
Patients with HNSCC with and without distant metastases had >3 tumour‐positive lymph nodes, and did not differ with respect to other risk factors. Statistical analysis was carried out using Student's t test, as well as statistical analysis of microarrays (SAM), to assess the false discovery rate for each gene. These analyses were supplemented with a newly developed method that computed deviations from gaussian‐order statistics (DEGOS). To validate the platform, normal mucosa of the head and neck was included as control.
Results
2963 genes were differently expressed between HNSCC and normal mucosa (t test; p<0.01). More rigorous statistical analysis with SAM confirmed the differential expression of most genes. The comparison of genes in HNSCC with and without metastases showed 150 differently expressed genes (t test; p<0.01), none of which, however, could be confirmed using SAM or DEGOS.
Conclusions
No evidence for a metastasis signature is found, and gene expression profiling of HNSCC has seemingly no value in determining the risk of developing distant metastases. The absence of such a signature can be understood when it is realised that, for HNSCC in contrast with breast cancer, the lymph nodes are a necessary in‐between station for haematogenous spread.
Head and neck squamous cell carcinoma (HNSCC) is the fifth most common type of carcinoma worldwide.1 Despite improvement in local control, survival has only marginally increased during the past three decades. A major negative factor in this respect is the development of metastatic disease at distant sites. Distant metastases occur in 10–20% of patients with HNSCC2,3,4,5 and the incidence may increase in the near future.6 Although new screening modalities have improved the detection rate of distant metastases at initial evaluation, these metastases continue to emerge during follow‐up in several patients. In all, 50% distant metastases are detected clinically within 9 months of treatment and 80% are detected within 2 years.7 The presence of distant metastases has dismal consequences for the patient. Adequate treatment is often not possible and life expectancy is dramatically decreased. It is important to better predict whether a patient will develop distant metastases during follow‐up, as this influences the decision on how to treat the patient for the initial HNSCC. In that case, unnecessary extensive treatment of the primary tumour can be omitted. The only option for this group of patients is effective local treatment in combination with adjuvant systemic treatment. Adjuvant systemic treatment, however, is still in the stage of development at present. To determine whether a patient will develop distant metastases during the course of the disease, an accurate marker that predicts distant metastases is urgently needed. Patients with multiple lymph node metastases, especially >3, have a relative high (up to 50%) risk of developing distant metastases.2,8,9,10 Unfortunately, this histopathological feature is still of little value for the individual patient, as <50% of these patients will develop distant metastases.
Over the past few years, gene expression profiling using microarray hybridisation, analysing thousands of genes simultaneously, has provided new insights into carcinogenesis and cancer dissemination. HNSCC has previously been studied with expression arrays by various authors as reviewed by Akervall,11 and in a few reports an association was shown between gene expression changes and clinically relevant variables, such as the presence of lymph node metastasis12 or patient survival.13,14. Chung et al14 reported a set of genes proposed to be predictive for lymph node metastases, which showed marked similarities with a gene set that was associated with metastatic disease in breast cancer.15 Recently, Roepman et al16 also identified a set of genes associated with the occurrence of lymph node metastases. Although an indication was found that this gene set holds promise of predicting the process of lymphatic metastatic disease, no analysis of survival, which is known to be strongly related to the development of distant metastases, was presented.
In breast cancer, van‘t Veer et al15 found an expression signature that correlated with the presence of distant metastases and that can possibly be applied to predict the occurrence of such a metastasis, although in a later study17 it was shown that the classifying gene set is strongly dependent on the way the data are analysed. Ramaswamy et al18 identified a gene expression signature of adenocarcinoma metastases (breast cancer, prostate cancer, medulloblastoma and large B cell lymphoma), which was present in some primary tumours and could therefore be used to predict which tumours have metastatic potential. On the basis of such studies, it was interpreted that some tumours already harbour a metastatic signature at an early stage, and that this persists in all or most cells in the primary tumour with metastatic potential during their lifetime.19 The implication is that, in some tumour types, such as breast cancer, cells with metastatic potential are able to directly disseminate from the primary tumour to distant sites, where they progress to overt metastases without previous passage through the lymph nodes.20 For HNSCC, the situation is possibly different. As there is a strong association between the presence of positive lymph nodes and the presence of distant metastases,2 it is conceivable that the lymph node is a necessary passing station for the ultimately haematogenously spreading cells.20 We analysed whether gene expression profiling can predict the development of distant metastases in patients with HNSCC with >3 lymph node metastases, a patient group with a 50% risk of developing distant metastases.8,9,10 To avoid the influence of confounding factors as much as possible, strict criteria were applied to the selection of patients with HNSCC. So, both groups consisted of patients with >3 tumour‐positive lymph nodes, and, to exclude the possibility that a negative result was due to the choice of platform or analytical methods, a panel of normal mucosal specimens was added.
Participants and methods
Patients and samples
The source of this study was a panel of liquid nitrogen‐stored HNSCC specimens that were collected during the past 10 years. We selected tumours from patients who either developed distant metastases during follow‐up (case group) or who remained disease free for a minimum follow‐up period of 3 years (control group; table 1). Further selection criteria were ⩾4 tumour‐positive lymph nodes, histologically tumour‐negative surgical margins, localisation in the oral cavity, oropharynx, hypopharynx or larynx, and no recurrent disease other than distant metastases. This selection was carried out to obtain a case–control with identical clinical features on a groupwide basis and a similar risk of developing distant metastases. The habits of smoking and drinking alcohol were scored, and, when information was available, tobacco use was calculated in pack‐years (number of packs per day, 25 cigarettes per pack, multiplied by years of active smoking) and alcohol consumption as unit‐years (glasses of beer, wine or liquor per day, multiplied by years of consumption).
Table 1 Patient and tumour characteristics.
| Patient code | Carcinoma | Patient | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Localisation | TNM | Extra‐capsular spread | No of tumour‐positive lymph nodes | Age at diagnosis (years) | Sex | Follow‐up | Smoking | Alcohol drinking | |||
| Yes/no | Pack‐years | Yes/no | Unit‐years | ||||||||
| M1 | Oral cavity | T3N2b | Yes | 5 | 71 | Male | DM lung, DOD | Yes | 25 | Yes | 400 |
| M2 | Oral cavity | T3N2c | Yes | 11 | 67 | Female | DM liver, DOD | Yes | Unknown | Yes | unknown |
| M3 | Larynx | T4N3 | Yes | 27 | 52 | Male | DM bone, DOD | Yes | 47 | Yes | 35 |
| M4 | Oral cavity | T4N2b | Yes | 7 | 65 | Male | DM lung, DOD | Yes | Unknown | Yes | Unknown |
| M5 | Oropharynx | T3N2b | Yes | 5 | 52 | Female | DM lung, DOD | Yes | 35 | Yes | 280 |
| M6 | Oral cavity | T2N2b | No | 7 | 48 | Male | DM pericardium, DOD | Yes | 30 | Yes | 40 |
| M7 | Oral cavity | T4N2b | No | 5 | 74 | Male | DM bone, DOD | Yes | 37 | No | 0 |
| M8 | Oral cavity | T4N2c | Yes | 5 | 58 | Male | DM liver, DOD | Yes | 50 | Yes | 400 |
| M9 | Oral cavity | T4N2c | Yes | 4 | 65 | Male | DM lung, bone, DOD | Yes | Unknown | Yes | Unknown |
| M10 | Hypopharynx | T1N3 | Yes | 5 | 54 | Male | DM bone, DOD | No | 0 | No | 0 |
| M11 | Oropharynx | T2N2b | Yes | 4 | 55 | Female | DM bone, liver, DOD | Yes | 30 | Yes | Unknown |
| NM1 | Larynx | T4N2b | Yes | 6 | 48 | Female | df 68 months | Yes | Unknown | Yes | Unknown |
| NM2 | Oropharynx | T2N2b | Yes | 8 | 48 | Male | df 44 months | Yes | 23 | Yes | 250 |
| NM3 | Oral cavity | T2N2b | Yes | 6 | 54 | Male | df 59 months | Yes | 48 | Yes | 560 |
| NM4 | Oropharynx | T2N2b | Yes | 4 | 52 | Male | df 67 months | Yes | 25 | Yes | 136 |
| NM5 | Oropharynx | T4N2b | Yes | 5 | 73 | Male | df 110 months | Yes | 26 | No | 0 |
| NM6 | Hypopharynx | T3N2c | Yes | 12 | 41 | Male | df 56 months | Yes | 10 | Yes | 150 |
| NM7 | Oral cavity | T4N2c | Yes | 6 | 49 | Female | df 58 months | Yes | 28 | Yes | 84 |
| NM8 | Oropharynx | T2N2c | No | 10 | 57 | Female | df 47 months | Yes | 40 | Yes | 400 |
df, disease free; DM, distant metastases; DOD, death from disease; HNSCC, head and neck squamous cell carcinoma; M, metastasised HNSCC; NM, non‐metastasised HNSCC; TNM, tumour‐node‐metastasis.
Several patients with tumours were not eligible because of lower risk nodal status (<4 positive lymph nodes), non‐surgical treatment, tumour‐positive surgical margins, distant metastases from other primary tumours or a too short disease‐free follow‐up period. Of the total 424 frozen tumours available for analysis, 11 were found eligible for the case group and eight for the control group; also eight normal mucosa specimens, obtained from the uvula of healthy controls without cancer were added for comparison and validation of the platform and analysis.
The Institutional Review Board of the VU University Medical Center, Amsterdam, The Netherlands, approved the study protocol, and written informed consent was obtained from all the patients.
RNA preparation
All samples were snap frozen in liquid nitrogen and stored at −80°C. Tumour percentage was estimated on 5‐μm‐thick sections stained with haematoxylin and eosin, with a mean tumour percentage of 62% in the case group (range 20–90%) and 60% in the control group (range 10–90%). In all, 15–20 30‐μm‐thick frozen sections were prepared with a cryomicrotome and carefully transferred to a chilled 1.5‐ml tube containing RNAbee (Campro Scientific, Veenendaal, The Netherlands) for intact RNA isolation according to the manufacturer's protocol. Quality control of total RNA samples was carried out with the RNA 6000 Pico LabChip kit (Agilent Technologies, Palo Alto, California, USA) and analysed on the Agilent 2100 Bioanalyzer. As a common reference for array hybridisation, the Universal Human Reference RNA from Stratagene (La Jolla, California, USA) was used.
Synthesis and labelling of cDNA
Because of possible non‐linear amplification of small amounts of RNA, we used non‐amplified total RNA for hybridisation on the arrays. The amounts of total RNA varied from 5 to 15 μg, depending on the size of the sample. Details of the preparation of labelled cDNA are provided in a previously published protocol.21 Cell samples were labelled with Cy3 (Fluorolink Cy3 Monofunctional Dye; Amersham, Freiburg, Germany) or Cy5 (Fluorolink Cy5; Amersham) for common reference.
Array hybridisation and scanning
The Human Release V.1.0 oligonucleotide library, containing 18 861 60‐mer oligonucleotides representing 17 260 unique genes as designed by Compugen (San José, California, USA) was obtained from Sigma‐Genosys (Zwijndrecht, The Netherlands). Hybridisation was carried out as previously described.21 Spots were quantified by Imagene V.5.5.4 software (Biodiscovery, Marina del Rey, California, USA), using the default settings. Local background was subtracted to obtain the signal mean. The expression platform we used has been described previously in detail,21 and a good correlation between array and Taqman results was obtained for several genes regarding the level of expression intensities.
Analysis
All expression intensities were transformed to log2 values and intensities <0 (below background) were classified as missing. Data were normalised by means of z score transformation.22 The expression intensity of each sample was calculated by subtracting the values of the Cy5 channel (reference) from those of the Cy3 channel (sample), yielding the Cy3:Cy5 ratio. The number of missing values varied per sample and had a mean of 13% of the values. To find potential classifying genes, differences in gene expression of HNSCCs with and without distant metastases were calculated with Student's t test (SPSS for Microsoft Windows). Only those genes with values of ⩾5 carcinomas per group were analysed. To confirm the findings, we additionally applied statistical analysis of microarrays (SAM) software V.1.21.23 A q value (%) and a false discovery rate value (%) was obtained for each gene. To validate our platform and analytical tools against existing data, we also compared the expression profiles of all HNSCC samples versus the normal mucosal samples.
A second analysis was carried out on the dataset according to the principles of ordered statistics. For each gene, the computed t value was converted to its exact gaussian analogue (z value) via the p value. All these z values were ranked and compared with the theoretical values from the gaussian distribution. A robust linear regression on the central 90% of the values gave rise to a tolerance region (with α = 1/n, where n is the number of genes considered). Genes were considered confirmed if they lay below the region for negative values of z and above for positive values of z. The analysis is based on deviations from gaussian‐order statistics (DEGOS). Details on DEGOS are available on request.
Annotation analysis was carried out with software available at http://source.stanford.edu. Cluster analysis of the latter comparison was carried out with the software program Spotfire DecisionSite (Spotfire, Somerville, MA, USA). Parameter settings were standard, with no filter or data adjustment, and the hierarchical unsupervised clustering was executed for genes and samples with Pearson's correlation and complete linkage selected.
Results
Characteristics of the study population
The average age in the case group (eight men and three women) was 60 (range 48–74) years, whereas the average age in the control group (five men and three women) was 53 (range 41–73) years. The groups did not differ with respect to tobacco and alcohol consumption, number of positive nodes and presence of extracapsular spread. Table 1 gives further details.
Expression profiles in HNSCC and normal mucosal samples
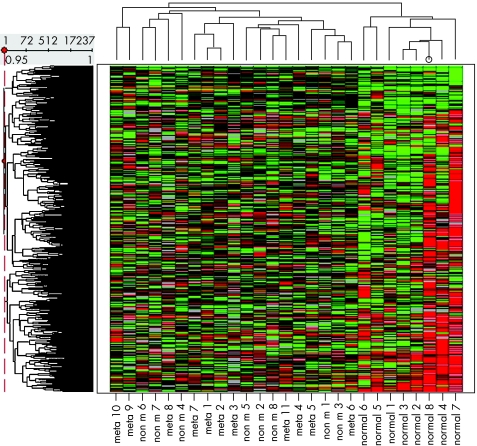
Nineteen HNSCC samples, with or without metastatic disease, and eight normal mucosa samples were compared with respect to the RNA expression profile. With unsupervised hierarchical clustering of all 17 237 genes in the 27 samples, two major groups were separated, the carcinomas and the normal mucosal samples (fig 1). We found 17.2% (2963/17 237) genes with a significantly different expression (p<0.01); 1063 of these were upregulated and 1900 downregulated in HNSCC. Table 2 shows the relationship between a certain cut‐off of the p value and the consequences for the number of different genes. Table 3 gives the list of the 50 genes showing the largest difference between these groups. A more extensive list of all differentially expressed genes can be found at http://www.jclinpath.com/supplemental.
Figure 1 Hierarchical clustering of 19 head and neck squamous cell carcinoma (HNSCC) specimens and eight normal mucosas of healthy controls. The dendrogram is based on the similarity of the 17 237 genes (left) and the 27 tissue samples (above). The clustering method involved complete linkage and correlation as the similarity measure. Empty values were replaced by the column average. The eight normal mucosas cluster as a separate group, as visualised at the right. non m, non‐metastasised HNSCC; meta, metastasised HNSCC.
Table 2 Relation between p value and number of differentially expressed genes.
| Upper cut‐off level p value | Differently expressed genes (n) | |
|---|---|---|
| Increased expression | Decreased expression | |
| HNSCC v normal mucosa | ||
| 0.01 | 1063 | 1900 |
| 0.001 | 563 | 931 |
| <0.001 | 310 | 483 |
| Metastasised v non‐metastasised HNSCC | ||
| 0.01 | 82 | 68 |
| 0.001 | 7 | 6 |
| <0.001 | 0 | 0 |
HNSCC, head and neck squamous cell carcinoma.
Table 3 Fifty most significantly different genes between normal mucosa and head and neck squamous cell carcinoma.
| Rank‐ number | HUGO‐ identifier | GenBank accession number | Description | Number | p‐ Value Student t | SAM | ||
|---|---|---|---|---|---|---|---|---|
| HNSCC | Normal mucosa | q Value (%) | Local fdr (%) | |||||
| Increased expression in HNSCC | ||||||||
| 1 | LOC492304 | AK025719 | Putative insulin‐like growth factor II‐associated protein | 19 | 7 | 1 | 0 | 0.26 |
| 2 | KIAA0261 | D87450 | KIAA0261 | 19 | 8 | 1 | 0 | 0 |
| 3 | HEIR1 | D28449 | Inhibitor of DNA binding 3 | 18 | 8 | 1 | 0 | 0 |
| 4 | C20orf3 | AB033767 | Chromosome 20 open reading frame 3 | 19 | 8 | 1 | 0 | 0 |
| 5 | Unknown | NM_005332 | Haemoglobin ζ | 19 | 8 | 1 | 0 | 0.23 |
| 6 | DDOST | NM_005216 | Dolichyl‐diphospho‐oligosaccharide‐protein glycosyltransferase | 19 | 8 | 1 | 0 | 0 |
| 7 | HBG2 | NM_000184 | Haemoglobin γ‐G | 17 | 8 | 1 | 0 | 0.32 |
| 8 | MSN | NM_002444 | Moesin | 19 | 8 | 1 | 0 | 0 |
| 9 | KIAA1922 | AF119868 | KIAA1922 protein | 19 | 8 | 1 | 0 | 0 |
| 10 | OPN | NM_000582 | Secreted phosphoprotein 1 (osteopontin) | 19 | 6 | 1 | 0 | 0 |
| 11 | LGALS1 | NM_002305 | Lectin, galactoside‐binding, soluble, 1 (galectin 1) | 18 | 8 | 1 | 0 | 0 |
| 12 | Unknown | NM_004052 | Data not found | 17 | 8 | 1 | 0 | 0 |
| 13 | IBP3 | NM_000598 | Insulin‐like growth factor binding protein 3 | 19 | 8 | 1 | 0 | 0 |
| 14 | KIAA0092 | NM_014679 | Translokin | 17 | 8 | 1 | 0 | 0 |
| 15 | MFHAS1 | NM_004225 | Malignant fibrous histiocytoma amplified sequence 1 | 19 | 8 | 1 | 0.09 | 1.55 |
| 16 | PIT1 | NM_005415 | Solute carrier family 20 (phosphate transporter) | 19 | 8 | 1 | 0 | 0 |
| 17 | HBGA | M11427 | Haemoglobin γ‐A | 16 | 8 | 1 | 0 | 0.16 |
| 18 | FNDC3B | AL157482 | Fibronectin type III domain containing 3B | 19 | 7 | 1 | 0 | 0 |
| 19 | NSUN5 | NM_018044 | NOL1/NOP2/Sun domain family, member 5 | 19 | 8 | 1 | 0.09 | 1.91 |
| 20 | IFI15 | NM_005101 | Interferon, α‐inducible protein (clone IFI‐15K) | 19 | 8 | 1 | 0 | 0 |
| 21 | H2B/H | NM_003523 | Histone 1, H2be | 19 | 8 | 1 | 0 | 0 |
| 22 | Unknown | Z36811 | Data not found | 19 | 8 | 1 | 0 | 0 |
| 23 | MCM6 | NM_005915 | MCM6 minichromosome maintenance | 19 | 8 | 1 | 0 | 0 |
| 24 | BST2 | NM_004335 | Bone marrow stromal cell antigen 2 | 19 | 8 | 1 | 0 | 0 |
| 25 | Unknown | K02847 | Data not found | 19 | 8 | 1 | 0 | 0 |
| Decreased expression in HNSCC | ||||||||
| 1 | THW | AF317550 | PERP, TP53 apoptosis effector | 19 | 8 | <0.001 | 0 | 0.06 |
| 2 | Unknown | AK000006 | Data not found | 19 | 8 | <0.001 | 0 | 0 |
| 3 | C1orf10 | NM_016190 | Chromosome 1 open reading frame 10 | 19 | 8 | <0.001 | 0 | 0 |
| 4 | PMI1 | NM_002435 | Mannose phosphate isomerase | 18 | 8 | <0.001 | 0 | 0.06 |
| 5 | CAGA | NM_002964 | S100 calcium binding protein A8 (calgranulin A) | 18 | 8 | <0.001 | 0 | 0.01 |
| 6 | ECM1 | NM_004425 | Extracellular matrix protein 1 | 19 | 8 | <0.001 | 0 | 0.10 |
| 7 | CL‐20 | NM_001423 | Epithelial membrane protein 1 | 17 | 8 | <0.001 | 0 | 0 |
| 8 | SERPINB2 | NM_002575 | Serine (or cysteine) proteinase inhibitor | 19 | 8 | <0.001 | 0 | 0.11 |
| 9 | HOP | AB019573 | Homeodomain‐only protein | 18 | 6 | <0.001 | 0 | 0 |
| 10 | CLCA4 | NM_012128 | Chloride channel, calcium activated, family member 4 | 14 | 6 | <0.001 | 0 | 0 |
| 11 | BICD1 | NM_001714 | Bicaudal D homologue 1 (Drosophila) | 18 | 8 | <0.001 | 0 | 0.10 |
| 12 | DKK1 | NM_012242 | Dickkopf homologue 1 (Xenopus laevis) | 19 | 8 | <0.001 | 0 | 0 |
| 13 | SPRR1B | NM_003125 | Small proline‐rich protein 1B (cornifin) | 14 | 8 | <0.001 | 0 | 0.01 |
| 14 | NAGK | NM_017567 | N‐acetylglucosamine kinase | 19 | 8 | <0.001 | 0 | 0 |
| 15 | VAV3 | NM_006113 | Vav 3 oncogene | 18 | 7 | <0.001 | 0 | 0.09 |
| 16 | DDX32 | NM_018180 | DEAH (Asp–Glu–Ala–His) box polypeptide 32 | 19 | 8 | <0.001 | 0 | 0 |
| 17 | BENE | U17077 | BENE protein | 19 | 8 | <0.001 | 0 | 0.07 |
| 18 | ZDHHC1 | U90653 | Zinc finger, DHHC domain containing 1 | 19 | 8 | <0.001 | 0 | 0 |
| 19 | ANXA1 | NM_000700 | Annexin A1 | 19 | 8 | <0.001 | 0 | 0 |
| 20 | Unknown | NM_015961 | Data not found | 18 | 8 | <0.001 | 0 | 0 |
| 21 | PAFAH | NM_005084 | Phospholipase A2, group VII | 19 | 8 | <0.001 | 0 | 0 |
| 22 | DAL1 | NM_012307 | Erythrocyte membrane protein band 4.1‐like 3 | 19 | 8 | <0.001 | 0 | 0 |
| 23 | M/NEI | M93056 | Serine (or cysteine) proteinase inhibitor | 17 | 8 | <0.001 | 0 | 0 |
| 24 | PADI1 | AB033768 | Peptidyl arginine deiminase, type I | 19 | 8 | <0.001 | 0 | 0.10 |
| 25 | SCEL | NM_003843 | Sciellin | 17 | 8 | <0.001 | 0 | 0.08 |
Nineteen HNSCC (11 with and eight without metastases) specimens were compared with 8 normal mucosa specimens using two‐sided Student's t test (a p value of 1 reflects a value between 0.999 and 1). Regarding SAM, the q value is shown, which is comparable to the p value of the t test. Local fdr is the false discovery rate in percentage. Further details on SAM can be found in the article by Cheadle et al22.
fdr, false discovery rate; HNSCC, head and neck squamous cell carcinoma; SAM, statistical analysis of microarrays.
SAM was carried out to assess the chance of a falsely positive gene identification. Table 3 shows details of the analysis of the 50 most different genes. In addition, of the top 100 downregulated genes, all had q value (comparable to the standard p value) <0.11%, and false discovery rate <0.1%. Two of the top 100 upregulated genes had a q value >2.5%, and 23 had a false discovery rate >5% (which was <15% for 22 of these).
Expression profiles in HNSCC with and without distant metastases
The expression profiles of 11 HNSCC specimens with and eight HNSCC specimens without distant metastatic disease were compared. When designing this study we took care to exclude all possible confounding factors. So the groups with and without distant metastases were at a similar group level with respect to sex, age, tumour‐node‐metastases stage, the number of tumour‐positive lymph nodes, and the smoking and alcohol drinking behaviour of the patients (table 1).
It was not possible to discriminate the metastasising tumours from the non‐metastasising ones by unsupervised clustering of all genes. Nevertheless, we found 150 of the 17 240 (0.8%; p<0.01) genes differently expressed on comparing profiles of carcinomas with and without distant metastases (additional information is available at http://www.jclinpath.com/supplemental); 82 genes showed a lower and 68 showed a higher expression in metastasised tumours. With lower p values, fewer genes were found to be different (table 2). Table 4 shows the gene sets with the most differential expression. We carried out additional analyses to exclude the possibility that a gene could be different simply as a result of chance. More rigorous testing was carried out using SAM, which resulted in finding no gene that fulfilled the program's criteria of being significantly different. q and false discovery rate values were >80%. This suggests the rejection of all genes uncovered by Student's t test owing to the high likelihood of being false positive. Also, another statistical approach (DEGOS) did not support the notion that the t test revealed truly different genes.
Table 4 Fifty most significantly different genes between metastasised and non‐metastasised head and neck squamous cell carcinoma.
| Rank number | HUGO identifier | GenBank accession number | Description | HNSCC (n) | p Value Student‘s t | |
|---|---|---|---|---|---|---|
| Non‐meta | Meta | |||||
| Downregulated in metastasised HNSCC | ||||||
| 1 | PCDH9 | AF085861 | Protocadherin 9 | 6 | 7 | <0.001 |
| 2 | GALR1 | NM_001480 | Galanin receptor 1 | 8 | 9 | <0.001 |
| 3 | COL4A4 | NM_000092 | Collagen, type IV, α4 | 8 | 10 | <0.001 |
| 4 | Unknown | U18909 | Data not found | 8 | 10 | <0.001 |
| 5 | TRIM62 | NM_018207 | Tripartite motif‐containing 62 | 8 | 9 | <0.001 |
| 6 | RERE | AK024214 | Arginine‐glutamic acid dipeptide (RE) repeats | 6 | 8 | <0.001 |
| 7 | Unknown | AF090927 | Data not found | 8 | 9 | <0.001 |
| 8 | SULT1A1 | NM_001055 | Sulfotransferase family, cytosolic, 1A, phenol‐preferring | 8 | 11 | <0.001 |
| 9 | AGTPBP1 | NM_015239 | ATP/GTP binding protein 1 | 6 | 11 | <0.001 |
| 10 | CHST5 | NM_012126 | Carbohydrate (N‐acetylglucosamine 6‐O) sulfotransferase 5 | 8 | 10 | <0.001 |
| 11 | PDPK1 | NM_002613 | 3‐Phosphoinositide dependent protein kinase‐1 | 6 | 8 | <0.001 |
| 12 | Unknown | AL137637 | mRNA; cDNA DKFZp434J035 (from clone DKFZp434J035) | 5 | 8 | <0.001 |
| 13 | TRAF5 | AB000509 | TNF receptor‐associated factor 5 | 8 | 10 | <0.001 |
| 14 | Unknown | NM_014097 | Data not found | 8 | 11 | |
| 15 | Unknown | AK025573 | Data not found | 5 | 6 | 0.001 |
| 16 | IDUA | NM_000203 | Iduronidase, α‐L‐ | 8 | 10 | 0.001 |
| 17 | TLR4 | NM_003266 | Toll‐like receptor 4 | 8 | 11 | 0.001 |
| 18 | MUC3A | M55406 | Mucin 3A, intestinal | 5 | 11 | 0.001 |
| 19 | Unknown | U48728 | Data not found | 6 | 11 | 0.001 |
| 20 | Unknown | NM_014684 | Data not found | 8 | 11 | 0.001 |
| 21 | DLL1 | NM_005618 | δ‐like 1 (Drosophila) | 6 | 11 | 0.002 |
| 22 | CCR9 | NM_006641 | Chemokine (C–C motif) receptor 9 | 7 | 10 | 0.002 |
| 23 | PLCE1 | NM_016341 | Phospholipase C, epsilon 1 | 8 | 10 | 0.002 |
| 24 | SEC22L1 | AK023270 | SEC22 vesicle trafficking protein‐like 1 (S cerevisiae) | 7 | 8 | 0.002 |
| 25 | Unknown | AF136408 | Data not found | 7 | 8 | 0.002 |
| Upregulated in metastasised HNSCC | ||||||
| 1 | DFNA5 | NM_004403 | Deafness, autosomal dominant 5 | 7 | 11 | 1 |
| 2 | MAP1B | NM_005909 | Microtubule‐associated protein 1B | 6 | 11 | 1 |
| 3 | PRNP | X82545 | Prion protein (p27‐30) (Creutzfeld‐Jakob disease) | 8 | 11 | 1 |
| 4 | RAB6A | AL049984 | RAB6A, member RAS oncogene family | 6 | 6 | 1 |
| 5 | VGCNL1 | AK002089 | Voltage gated channel like 1 | 8 | 10 | 1 |
| 6 | Unknown | AK022068 | CDNA FLJ12006 fis, clone HEMBB1001585 | 5 | 11 | 1 |
| 7 | GARP | NM_005512 | Leucine rich repeat containing 32 | 7 | 11 | 1 |
| 8 | FLJ20313 | NM_017762 | Myotubularin‐related protein 10 | 8 | 11 | 1 |
| 9 | FLJ20397 | NM_017802 | Hypothetical protein FLJ20397 | 8 | 10 | 0.999 |
| 10 | MEFV | NM_000243 | Mediterranean fever | 8 | 11 | 0.999 |
| 11 | PMAIP1 | D90070 | Phorbol‐12‐myristate‐13‐acetate‐induced protein 1 | 8 | 11 | 0.999 |
| 12 | UBE2I | NM_003345 | Ubiquitin‐conjugating enzyme E2I (UBC9 homolog, yeast) | 8 | 11 | 0.999 |
| 13 | RPS2 | NM_016281 | Ribosomal protein S2 | 8 | 11 | 0.999 |
| 14 | DPYSL3 | NM_001387 | Dihydropyrimidinase‐like 3 | 7 | 11 | 0.999 |
| 15 | SNX5 | NM_014426 | Sorting nexin 5 | 8 | 11 | 0.999 |
| 16 | FLJ12666 | AK022728 | Chromosome 1 open reading frame 108 | 8 | 11 | 0.999 |
| 17 | FLJ34870 | AK022384 | FLJ34870 protein | 7 | 5 | 0.999 |
| 18 | PCDH7 | NM_002589 | BH‐protocadherin (brain‐heart) | 8 | 11 | 0.999 |
| 19 | KIAA1036 | NM_014909 | KIAA1036 | 7 | 9 | 0.999 |
| 20 | GNA12 | NM_007353 | Guanine nucleotide binding protein (G protein) α 12 | 8 | 11 | 0.999 |
| 21 | FTSJ1 | NM_012280 | FtsJ homologue 1 (E coli) | 8 | 10 | 0.999 |
| 22 | SAV1 | AK023071 | Salvador homolog 1 (Drosophila) | 8 | 11 | 0.999 |
| 23 | RANBP17 | L08438 | RAN binding protein 17 | 5 | 9 | 0.999 |
| 24 | UBA2 | NM_005499 | SUMO‐1 activating enzyme subunit 2 | 8 | 11 | 0.999 |
| 25 | SERPINA1 | M26123 | Serine (or cysteine) proteinase inhibitor | 8 | 11 | 0.999 |
In all, 11 HNSCC specimens with and eight without metastases were compared using two‐sided Student's t test (a p value of 1 reflects a value between 0.999 and 1). None of the genes could be confirmed using SAM.22
HNSCC, head and neck squamous cell carcinoma; SAM, statistical analysis of microarrays.
Discussion
HNSCC shows heterogeneity with respect to metastatic behaviour, and the current set of clinical markers is not sufficiently accurate to predict which patient is most at risk of distant metastases. This study was designed to find a set of genes that was differently expressed between metastasising and non‐metastasising HNSCCs. Patients were carefully selected for the presence of >3 lymph node metastases, a factor associated with a relatively high risk of distant metastases. When lymph node metastases in the neck are diagnosed, the chance of survival is halved. We also initially tried to select a case and a control group without lymph node metastases, but we could not find a single case in 424 tumours, strongly supporting the importance of the lymph node compartment for metastasising HNSCC.
To exclude that our platform or our analytical methods might influence the outcome, we compared the expression profile in normal and tumour tissue and discovered many genes to be differently expressed. We have purposely chosen normal tissue from patients without cancer as tumour‐adjacent normal tissue bears the risk of being genetically aberrant.24 Several genes were found to be different between cancerous and normal tissue, and this list includes those genes associated with signal transduction, cell structure, cell cycle, transcription, cell–cell adhesion, cell–matrix interaction and apoptosis. Other reports also found differentially expressed genes, although with lower numbers.25,26,27,28,29,30 Some highly different genes are shared between these reports (eg MSN, SCEL, SPARC, collagens and cytokeratins), and, in addition, similar cellular processes have been reported to be associated. This present set of genes with differential expression has a relatively strong effect, as unsupervised clustering of all available genes (filtered for expression level) generated a dendrogram that separated out the normal tissues and the carcinomas. With more stringent methods such as SAM and DEGOS, most of the highly differentially expressed genes could be confirmed. Thus, it is possible with the presently used expression platform and analysis to generate relevant information, in line with previously published data.
We have found a panel of 150 genes that had a differential expression between tumours with >3 lymph node metastases that either did or did not give rise to distant metastases (p<0.01 with Student's t test), but this test is not optimal as it does not exclude the possibility that these differences have occurred by chance. This number of 150 differentially expressed genes is roughly what would be expected, if randomly distributed values for each gene were assumed. We identified no relevant gene using SAM, an established and rather stringent method that uses permutations to increase the power of significance analysis. Either a gene was not different or there was a high likelihood of it being falsely different. A second method, DEGOS, which was recently developed in our centre to overcome some intrinsic analysis problems with SAM, also did not provide evidence that the genes were unambiguously differentially expressed. Our study does not provide evidence for a metastatic signature, and indicates that expression profiling has seemingly no additional value in predicting the development of distant metastases. The current method to assess the risk of distant metastases, the examination of lymph nodes for the presence of cancer, unfortunately cannot be improved.
Importantly, relatively small patient groups have been used in our study. Nevertheless, when comparing the eight normal mucosas with eight randomly chosen HNSCC specimens, SAM was able to find most of the genes that were also different when data of all 19 HNSCCs were used. This indicates that a large difference in expression is hardly influenced by the number of specimens of available in this study. Also when studying breast cancer, we were able to find a metastatic signature, even when only seven metastatic tumours were compared with seven non‐metastatic carcinomas.31 Our results are in line with those of Cromer et al26 who studied 15 metastasising and 11 non‐metastasising hypopharyngeal carcinomas. Although these authors report a signature of 164 differentially expressed genes, they also concluded that it was too early to state whether a useful signature exists. There is the possibility that analysis of much larger numbers of patients with HNSCC will lead to the discovery of a distant metastases expression signature. Its existence will probably be based on several genes that show a small difference in expression between metastasising and non‐metastasising HNSCC, or it will be valid only on a subgroup of HNSCC. If a signature with such characteristics exists, it is doubtful whether it will have much value for the individual patient.
We have studied tumours from various locations in the head and neck area, and there was a relative over‐representation of tumours of the oral cavity in the metastasising group of tumours. It is unclear at this moment whether the expression profile of HNSCCs differs between subsites and whether this influenced the outcome of our study. The numbers of tumours analysed in this study are too small to correct for this potential bias and for the fact that there was no subsite categorisation of the HNSCC after global unsupervised clustering using the information of all genes (fig 1).
In conclusion, we have used microarray expression analysis and explored its potential for diagnostic purposes in HNSCC. No evidence for a distant metastasis signature was found, indicating that expression profiling has seemingly no value in predicting the development of distant metastases. HNSCC may differ in this respect from other tumour types such as breast and prostate cancer. The fact that lymph nodes are a necessary in‐between station for haematogenous spread may explain the absence of a distant metastasis‐associated HNSCC expression profile.
Supplementary Material
Acknowledgements
We thank Wim de Boer for analytical support, and Paul Eijk and Paul van den IJssel for printing slides and technical and analytical assistance.
Abbreviations
DEGOS - deviations from gaussian‐order statistics
HNSCC - head and neck squamous cell carcinoma
SAM - statistical analysis of microarrays
Footnotes
This project is financially supported by the European Union FP6, LHSC‐CT‐2005‐018911, DISMAL. The publication reflects only the authors' view. The European Commission is not liable for any use that may be made of the information obtained.
Competing interests: None.
References
- 1.Pisani P, Bray F, Parkin D M. Estimates of the world‐wide prevalence of cancer for 25 sites in the adult population. Int J Cancer 20029772–81. [DOI] [PubMed] [Google Scholar]
- 2.Leemans C R, Tiwari R, Nauta J J.et al Regional lymph node involvement and its significance in the development of distant metastases in head and neck carcinoma. Cancer 199371452–456. [DOI] [PubMed] [Google Scholar]
- 3.Al‐Othman M O, Morris C G, Hinerman R W.et al Distant metastases after definitive radiotherapy for squamous cell carcinoma of the head and neck. Head Neck 200325629–633. [DOI] [PubMed] [Google Scholar]
- 4.Gourin C G, Johnson J T. Surgical treatment of squamous cell carcinoma of the base of tongue. Head Neck 200123653–660. [DOI] [PubMed] [Google Scholar]
- 5.Harrison L B, Lee H J, Pfister D G.et al Long term results of primary radiotherapy with/without neck dissection for squamous cell cancer of the base of tongue. Head Neck 199820668–673. [DOI] [PubMed] [Google Scholar]
- 6.Taneja C, Allen H, Koness R J.et al Changing patterns of failure of head and neck cancer. Arch Otolaryngol Head Neck Surg 2002128324–327. [DOI] [PubMed] [Google Scholar]
- 7.Merino O R, Lindberg R D, Fletcher G H. An analysis of distant metastases from squamous cell carcinoma of the upper respiratory and digestive tracts. Cancer 197740145–151. [DOI] [PubMed] [Google Scholar]
- 8.de Bree R, Deurloo E E, Snow G B.et al Screening for distant metastases in patients with head and neck cancer. Laryngoscope 2000110397–401. [DOI] [PubMed] [Google Scholar]
- 9.Vandenbrouck C, Eschwege F, De la Rochefordiere A.et al Squamous cell carcinoma of the pyriform sinus: retrospective study of 351 cases treated at the Institut Gustave‐Roussy. Head Neck 1987104–13. [DOI] [PubMed] [Google Scholar]
- 10.Alvi A, Johnson J T. Development of distant metastasis after treatment of advanced‐stage head and neck cancer. Head Neck 199719500–505. [DOI] [PubMed] [Google Scholar]
- 11.Akervall J. Gene profiling in squamous cell carcinoma of the head and neck. Cancer Metastasis Rev 20052487–94. [DOI] [PubMed] [Google Scholar]
- 12.Nagata M, Fujita H, Ida H.et al Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer 2003106683–689. [DOI] [PubMed] [Google Scholar]
- 13.Belbin T J, Singh B, Barber I.et al Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res 2002621184–1190. [PubMed] [Google Scholar]
- 14.Chung C H, Parker J S, Karaca G.et al Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 20045489–500. [DOI] [PubMed] [Google Scholar]
- 15.van't Veer L J, Dai H, van de Vijver M J.et al Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002415530–536. [DOI] [PubMed] [Google Scholar]
- 16.Roepman P, Wessels L F, Kettelarij N.et al An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 200537182–186. [DOI] [PubMed] [Google Scholar]
- 17.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays. Lancet 20053651684–1685. [DOI] [PubMed] [Google Scholar]
- 18.Ramaswamy S, Tamayo P, Rifkin R.et al Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA 20019815149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernards R, Weinberg R A. A progression puzzle. Nature 2002418823. [DOI] [PubMed] [Google Scholar]
- 20.Pantel K, Brakenhoff R H. Dissecting the metastatic cascade. Nat Rev Cancer 20044448–456. [DOI] [PubMed] [Google Scholar]
- 21.Buermans H P, Redout E M, Schiel A E.et al Microarray analysis reveals pivotal divergent mRNA expression profiles early in the development of either compensated ventricular hypertrophy or heart failure. Physiol Genomics 200521314–323. [DOI] [PubMed] [Google Scholar]
- 22.Cheadle C, Vawter M P, Freed W J.et al Analysis of microarray data using Z score transformation. J Mol Diagn 2003573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher V G, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001985116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braakhuis B J, Leemans C R, Brakenhoff R H. Using tissue adjacent to carcinoma as a normal control: an obvious but questionable practice. J Pathol 2004203620–621. [DOI] [PubMed] [Google Scholar]
- 25.Mendez E, Cheng C, Farwell D G.et al Transcriptional expression profiles of oral squamous cell carcinomas. Cancer 2002951482–1494. [DOI] [PubMed] [Google Scholar]
- 26.Cromer A, Carles A, Millon R.et al Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene 2004232484–2498. [DOI] [PubMed] [Google Scholar]
- 27.Al Moustafa A E, Alaoui‐Jamali M A, Batist G.et al Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene 2002212634–2640. [DOI] [PubMed] [Google Scholar]
- 28.Ha P K, Benoit N E, Yochem R.et al A transcriptional progression model for head and neck cancer. Clin Cancer Res 200393058–3064. [PubMed] [Google Scholar]
- 29.Alevizos I, Mahadevappa M, Zhang X.et al Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 2001206196–6204. [DOI] [PubMed] [Google Scholar]
- 30.El‐Naggar A K, Kim H W, Clayman G L.et al Differential expression profiling of head and neck squamous carcinoma: significance in their phenotypic and biological classification. Oncogene 2002218206–8219. [DOI] [PubMed] [Google Scholar]
- 31.Woelfle U, Cloos J, Sauter G.et al Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res 2003635679–5684. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.