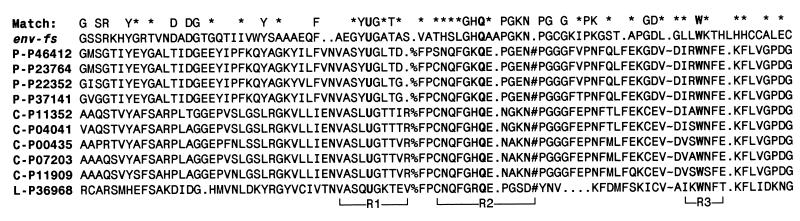
Figure 1.
Sequence alignment of HIV-1 env-fs sequence vs. Se-dependent GPx. Sequences are single-letter amino acid code, with U as the symbol for Sec, encoded by the UGA codon in RNA. GPx sequences are listed by Swiss-Prot database accession number, after a prefix of either P- (plasma), C- (cellular), or L- (phospholipid hydroperoxide), identifying three major families of Se-dependent GPx sequences. The N terminal of mature GPx protein begins at the fourth residue in the alignment. Env-fs amino acids identical to one or more of the aligned GPx sequences are shown as letters in the “match” line above the alignment; similar residues are indicated by an asterisk. Three active-site regions that are adjacent in 3-D space are shown below the alignment, labeled R1–R3. All three regions and their essential catalytic amino acids, Sec, Gln, and Trp (U, Q, and W, respectively, shown highlighted in bold), are represented in the truncated env-fs sequence. There are three large internal deletions in env-fs relative to the GPx sequences (numbered 1–3 in Fig. 2), at the locations indicated by the symbols %, #, and ∼ in the alignment, involving 19, 11, and 41 residues, respectively. As computed by using the alignment shown, i.e., omitting the three internal GPx regions where there are major deletions in the putative HIV-1 GPx homolog, the total similarity score of the env-fs sequence to the aligned GPx sequences is 6.3 SD above the average similarity score computed for 100 optimally aligned randomly shuffled sequences of identical composition. This significance score rises to 6.7 SD if the comparison is made only to the plasma GPx sequences, to which the HIV sequence is most similar overall; however, it also has features of the cellular and phospholipid hydroperoxide types of GPx.