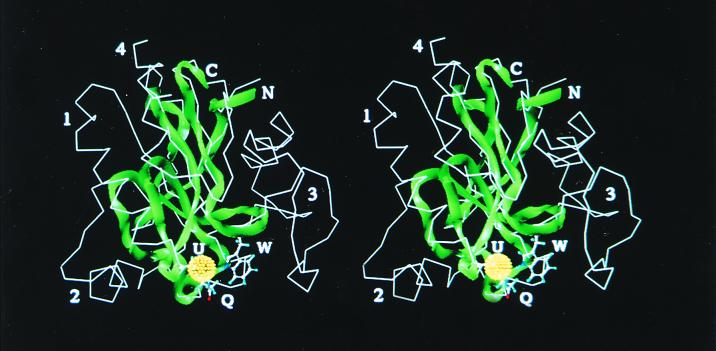
Figure 2.
Stereo view of the aligned protein backbones of the putative HIV-1 GPx homology model (ribbon rendition) and the x-ray crystal structure of the bovine GPx monomer (line rendition). The active site is in the lower foreground, with the side chains of the catalytic triad (Sec, Gln, and Trp) shown in a ball and stick rendition and labeled U, Q, and W, respectively; the Se atom is shown as a dot sphere. The C and N terminals of the HIV-1 peptide are labeled. The regions labeled by numbers 1–3 correspond to the major internal deletions shown as %, #, and ∼ in the sequence alignment of Fig. 1. The region labeled 4 is a C-terminal deletion, not shown in the alignment. These four deletions are domains of the bovine GPx that have no equivalent in the highly truncated HIV-1 GPx homolog. The deleted regions include a helix involved in dimer formation (no. 2) and another region (no. 3) that is involved in dimer and tetramer formation. No. 1 is an internal deletion between the U and the Q. Despite these deletions, the structural and catalytic core of the enzyme is conserved in the truncated HIV-1 homolog.