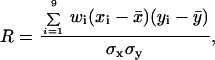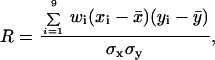Patterns of correlation between the presence or absence of NCp7 for strand transfer or for the pausing profile. (
A) The frequency of strand transfer yielded by HIV-1 RT on naked RNA is plotted against the one found on a NCp7/RNA complex. The values are given as frequency of recombination per nucleotide and multiplied by 10
4. The correlation coefficient
R between two sets of conditions (where the local rates are, respectively,
xi and
yi) is computed as described in ref.
26:
where
x̄ and
ȳ are the average rates,
wi is the statistical weight attributed to each interval (relative to its size in nucleotides as given in Table
1), and σ
x and σ
y are the standard deviations as tabulated in row 11 of Table
1. The calculation of the regression line was also performed as described (
26). The low value of
R indicates a nonsignificant correlation. (
B) Extension of a labeled primer in the presence (+) or absence (−) of NCp7 with HIV-1 RT. The reaction was performed under the same experimental conditions as the recombination assay (also see
Materials and Methods), and the incubation times are given above each lane of the gel. The boxes (
Left) show the position of the intervals of the region of homology. K, sequence coding for the resistance to kanamycin (also see Fig.
1). (
C) After determination of the strength of the stops during reverse transcription as described in
Materials and Methods, the corresponding values found for HIV-1 RT on naked RNA were plotted against those found on a NCp7/RNA complex. Only the pauses of a strength higher than 0.1 for at least one of the two experimental conditions tested are shown. The correlation coefficient
R is close to 1.00.