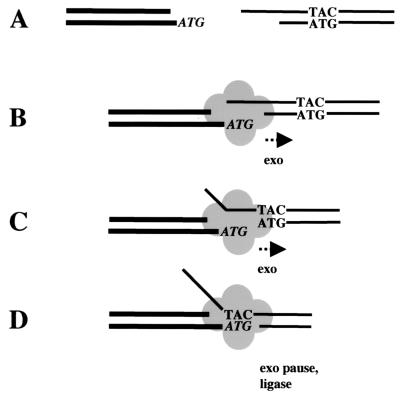
Figure 5.
A hypothetical model of microhomology searching by Mre11. (A) Broken DNA ends are generated that contain a sequence identity (“ATG”) in the single-stranded overhang of one end (thick lines) that is also present in the double-stranded region of the other end (thin lines). (B) A putative multimer of Mre11 binds both DNA ends simultaneously. The 5′ overhangs do not match, thus Mre11 exonuclease activity is stimulated on one end (here shown as the end on the right). (C) After excision of a few nucleotides, the overhangs still cannot base pair with one another, so Mre11 exonuclease activity proceeds further. (D) Degradation of the ATG present on the bottom strand of the right-hand DNA molecule uncovers the TAC on the top strand, thus creating cohesive ends and pausing exonuclease activity, which increases the probability of a DNA ligase sealing the nick on the bottom strand. Other enzymes would then be necessary to repair the top strand.