Abstract
Objective
One of the factors leading to an invasive phenotype is the nm23 family of metastases‐associated genes. Of the six known members, nm23‐H1 is the most frequently studied potential anti‐metastatic gene in cervical cancer. However, the possible molecular links to oncogenic human papillomavirus (HPV) are completely unexplored as yet.
Materials and methods
As a part of the HPV‐Pathogen Istituto Superiore di Sanità study, a series of 150 squamous cell carcinomas (SCCs) and 152 cervical intraepithelial neoplasia (CIN) lesions were examined by immunohistochemical staining for nm23‐H1, and tested for HPV by polymerase chain reaction (PCR) with three sets of primers (MY09/11, GP5+/GP6+ and short PCR fragment). Follow‐up data were available on all patients with SCC, and 67 CIN lesions were monitored by serial PCR for clearance or persistence of HPV after cone treatment.
Results
A linear decrease (p = 0.001) was observed in nm23‐H1 expression, starting from CIN1 (85% with normal expression), with the most dramatic down regulation on transition from CIN2 (70% normal) to CIN3 (39%) and further to SCC (25%). Reduced expression was associated with CIN3 or cancer at an odds ratio 8.72 (95% confidence interval 4.13 to 18.41). Nm23‐H1 was of no use as a marker of the high‐risk human papillomavirus (HR‐HPV) type, and it did not predict clearance or persistence of HR‐HPV after treatment of CIN. Importantly, nm23‐H1 expression was a significant prognostic factor in cervical cancer, reduced expression being associated with lower survival (p = 0.022) in univariate analysis. In the multivariate (Cox) regression model, however, only the International Federation of Gynecology and Obstetrics stage (p = 0.001) and age (p = 0.011) remained independent prognostic predictors.
Conclusions
Down‐regulated nm23‐H1 expression is markedly associated with progression from CIN2 to CIN3, and predicts poor prognosis in cervical cancer. Nm23‐H1 down regulation is probably orchestrated by mechanisms independent of HR‐HPV oncoproteins and is possibly related to the emergence of a proteolytic phenotype.
The high‐risk human papillomavirus (HR‐HPV) types are associated with cervical cancer in almost 100% of cases, in contrast with the low‐risk HPV (LR‐HPV) types that are rarely found in cervical cancer and its precursors.1,2,3,4,5,6 The different oncogenic potentials of LR‐HPV and HR‐HPV seem to be linked, at least in part, to the functions of two viral oncogenes, E6 and E7, and their different interactions with the key regulatory proteins of the cell cycle, p53 and Prb.2,3,4,5,7,8 Although the E6 oncoprotein of HR‐HPV (but not LR‐HPV) initiates degradation of the p53 tumour suppressor protein, HPV E7 of HR‐HPV (but not LR‐HPV) binds to pRB, resulting in G1–S transition of the cell cycle.2,3,4,7,8,9 These HPV‐type‐dependent functions of E6 and E7 are in contrast with those of another HPV oncoprotein E5, which activates potent nuclear transcription factors (c‐fos, myc, Ets1, Ets2, Elk‐1, c‐jun) through the extracellular signal‐regulated kinase (ERK)–mitogen‐activated protein kinase signalling pathway, but in an HPV‐type‐independent manner, as suggested by the fact that ERK1 expression is completely unrelated to types of HR‐HPV.10
Apart from angiogenesis,11 the development of an invasive phenotype from cervical cancer precursors (cervical intraepithelial neoplasia (CIN)) has been recently ascribed to a group of proteins encoded by the nm23 family of metastases‐associated genes.12,13 The human nm23 gene family includes several members (nm23‐H1, nm23‐H2, DR‐nm23, nm23‐H4, nm23‐H5 and nm23‐H6),14 with particular interest focused on nm23‐H1. Since its characterisation, nm23‐H1 has been extensively studied as a potential anti‐metastatic gene in several human tumours,15,16,17,18,19 including cervical cancer.12,13,20,21,22,23,24,25 The products of the nm23‐H1 and nm23‐H2 genes are identical to human nucleoside diphosphate (NDP) kinases A and B, respectively, both catalysing the phosphorylation of nucleoside diphosphates to nucleoside triphosphates.14,26 Evidence suggests that the nm23 protein can function in the transciptional regulation of c‐myc expression.26
In cervical cancer and its precursor lesions, nm23‐H1 has been analysed in a sizable number of studies,12,13,21,22,23,24,25,27,28,29,30,31 but the results are conflicting. Although some studies have established a definite anti‐metastatic effect for nm23‐H1,24,28,29 others failed to do so,12,13 or the number of cases was too small to draw definite conclusions. Regarding CIN lesions, some studies report an inverse relationship between nm23‐H1 expression and CIN grade,23 whereas others report increased nm23‐H1 expression with increased high‐grade CIN.12,13,25 Similarly, the prognostic value of nm23‐H1 in cervical cancer remains a highly controversial issue.12,13,20,21,22,24,27,28,30,31,32 Unexpectedly, none of these studies have assessed nm23‐H1 expression in relation to HPV in cervical lesions, and no in vitro studies are available on the possible interactions between HPV and the nm23 gene family either.
As a part of our HPV‐Pathogen Istituto Superiore di Sanità (ISS) study devoted to a systematic search for new potential biomarkers in HPV‐related cervical carcinogenesis,8,10,11,33 we analysed a series of cervical carcinomas and CIN lesions to assess whether nm23‐H1 expression may be of any use in predicting several of the intermediate endpoint markers of cervical cancer—namely (a) the grade of CIN; (b) the type of HR‐HPV; (c) clearance of the virus after eradication of CIN; or (d) prognosis of cervical cancer. Expression of nm23‐H1 was studied in CIN lesions treated by conisation and monitored by serial polymerase chain reaction (PCR) assays for clearance or persistence of HPV, and survival data of patients with cervical cancer were related to nm23‐H1 expression in surgical samples.
Materials and methods
Materials
The materials of this study comprise the retrospective component of the HPV‐Pathogen ISS project,34 and were collected from the files of the pathology departments of two Italian hospitals (S Orsola Malpighi Hospital, Bologna, Italy and Maggiore Hospital, University of Trieste, Trieste, Italy). Altogether, this prospective biopsy material was taken from 302 patients with either invasive cervical squamous cell carcinoma (SCC) or CIN diagnosed and treated in these two hospitals between 1986 and 2002. Of these 302 patients, 114 patients with CIN and 38 patients with SCC were from S Orsola Malpighi Hospital, and 38 patients with CIN and 112 with SCC were from Maggiore Hospital. The mean age of all patients with CIN was 35.5 (range 18–79) years and that of patients with SCC was 59.2 (range 27–89) years (p = 0.001).
Available data
All the patients from S Orsola Malpighi Hospital had their HPV status determined by PCR, as reported before,35,36,37 whereas the patients from Maggiore Hospital were tested for HPV in our study. Complete follow‐up data were available on all 150 patients with SCC, with a mean follow‐up of 51.7 (range 1–218) months. Furthermore, all patients with CIN from S Orsola Malpighi Hospital had been followed up at 6‐month intervals after cone treatment (mean 10.5, range 2.4–27.6 months), and subjected to repeated colposcopy, cervical smear test and biopsy (if residual suspected). A minimum of two (up to seven) serial PCR analyses were available from 67 patients and recently reported in a study on HPV clearance.37
Methods
Biopsy
Both the colposcopic biopsy samples and surgical samples were fixed in 10% buffered formalin, embedded in paraffin wax and processed to obtain 5‐μm‐thick sections that were stained with haematoxylin–eosin for routine diagnosis. All slides were re‐examined to confirm the diagnosis. On histological examination, the lesions were graded using the CIN nomenclature and categorised as CIN1, CIN2 and CIN3. The histological diagnosis of SCC was confirmed in all cases, and two adenocarcinomas present in the original cohort were excluded from this series.
Immunohistochemistry for nm23‐H1
Immunohistochemical (IHC) staining for the NDP kinase nm23‐H1 (also known as NME1) expression was completed following standard IHC procedures. Briefly, 5‐μm‐thick sections of paraffin wax cut on poly‐l‐lysine‐coated microscopy slides were deparaffinised and rehydrated in graded alcohols. The sections were heated in citrate buffer (0.01 mol/l, pH 6.0, Dako Target Retrieval Solution, Dako (Carpinteria, California, USA)) in a microwave oven (85–95°C, 3×5 min), followed by blocking the non‐specific binding sites with normal rabbit serum. Sections were incubated with the primary antibody, polyclonal rabbit antihuman nm23 protein (Catalogue #A0096, DakoCytomation Denmark A/S, Glostrup, Denmark; dilution 1:50), in a humidified chamber for 1 h at room temperature. This affinity‐isolated antibody was isolated using an immobilised peptide from the nm23 protein, which corresponds to a carefully selected synthetic peptide from the human nm23 protein used for immunisation. The antibody reacts with nm23‐H1 and nm23‐H2 gene products. Incubation with the primary antibody was followed by incubation with the biotinylated secondary antibody, polyclonal goat antirabbit immunoglobulin G (#6720, Abcam, dilution 1:200 (Abcam, Cambridge, UK)). Slides were then processed with universal LSAB‐2 single reagents (peroxidase) kit (DakoCytomation), and expression of nm23‐H1 was localised by incubation with diaminobenzidine. As a final step, the slides were stained with light haematoxylin counterstaining. Negative controls were similarly processed by omitting the primary antibody, and biopsy samples from breast cancer were used as positive controls.
Evaluation of IHC staining
IHC staining was examined with a light microscope (Leitz Diaplan, Leitz Wetzlar, Germany), equipped with a digital camera (Leica DG300). In normal squamous epithelium, nm23‐H1 staining was universally strong throughout the cervix. Nm23‐H1 staining was mainly cytoplasmic, but some nuclear staining was also detectable. In the original grading of IHC staining, a semiquantitative scoring into 4 categories was used: 0, negative (no nm23‐H1 staining); 1, weak staining (markedly reduced nm23‐H1 expression, with major negative areas or with a diffuse weak staining); 2, moderate staining (slightly to moderately reduced nm23‐H1, with minor areas without staining or a diffuse staining weaker than normal); and 3, strong staining (equivalent to staining of the normal epithelium; figs 1–5). In statistical analysis, the staining results were treated as dichotomous categorical variables (strong v reduced), or using the four‐tier categorisation.
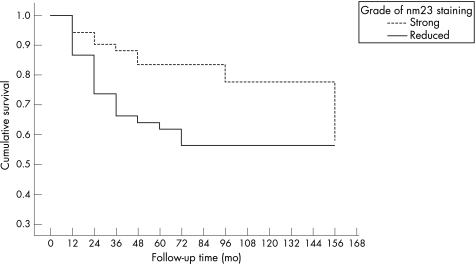
Figure 1 Cumulative survival related to nm23‐H1 expression in Kaplan–Meier analysis. mo, months.
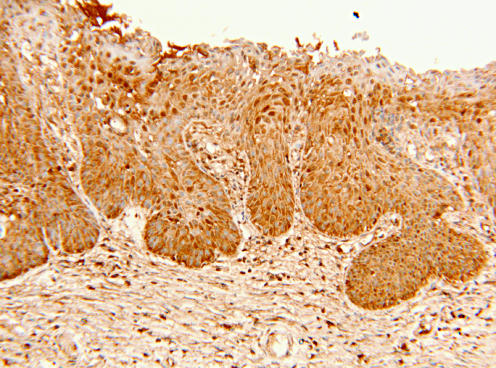
Figure 2 Normal transformation zone showing the process of immature squamous metaplasia. Also, the metaplastic squamous epithelium shows intense expression of nm23‐H1 protein in all layers of the epithelium (immunohistochemistry for nm23‐H1, original magnification ×100).
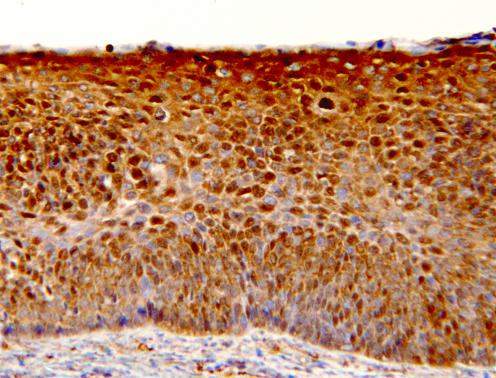
Figure 3 A low‐grade cervical intraepithelial neoplasia 1 lesion, showing intense staining for nm23‐H1 throughout the full thickness of the lesion, being mainly cytoplasmic; many positive nuclei are also present. The underlying stroma is nm23‐H1 negative (immunohistochemistry for nm23‐H1, original magnification ×200).
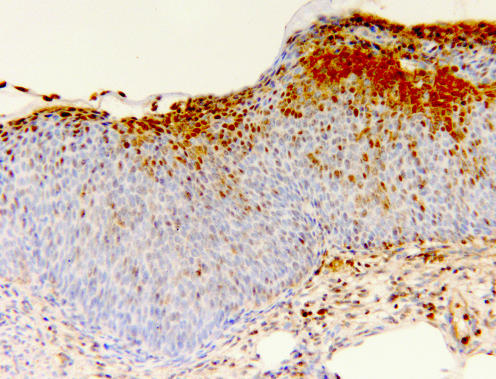
Figure 4 A high‐grade cervical intraepithelial neoplasia 3 lesion, with markedly reduced nm23‐H1 expression. Positive staining is confined to the uppermost layers of the epithelium, whereas the lower two thirds remains entirely nm23‐H1 negative, except for some single scattered positive cells (immunohistochemistry for nm23‐H1, original magnification ×100).
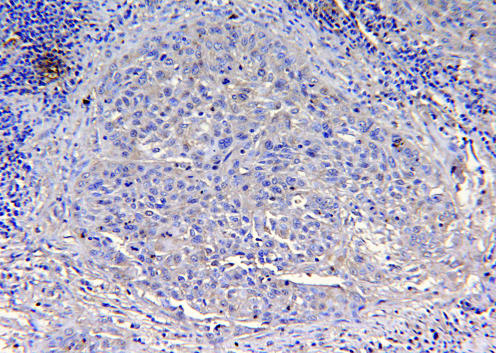
Figure 5 Details of an invasive squamous cell carcinoma. In contrast with the intense diffuse staining seen in cervical intraepithelial neoplasia 1 lesions, only a weak cytoplasmic staining is present in some single cancer cells, making it difficult to discern the invasive focus from the surrounding stroma. This represents a typical case of markedly reduced nm23‐H1 expression (immunohistochemistry for nm23‐H1, original magnification ×100).
HPV testing
The patients with CIN (n = 114) and SCC (n = 38) from S Orsola Malpighi Hospital had already been tested for HPV for other purposes by PCR, as recently reported.35,36,37 In our study, the 150 sections embedded in paraffin wax (112 SCCs and 38 CINs) from Maggiore Hospital were subjected to HPV testing by PCR.
Polymerase chain reaction
To verify the extraction and the quality of DNA from the tissues embedded with paraffin wax, 5 μl of each sample was amplified with a primer set recognising the β‐actin gene (sense 5′‐GGCGGCACCACCATGTACCCT‐3′, anti‐sense 5′‐AGGGGCCGGACTCGTCATACT‐3′). The PCR mix contained 200 μM of each deoxyribonucleotide triphosphate, 1.5 mM magnesium chloride (MgCl2), 1× PCR buffer, 40 pmol sense and anti‐sense primer, and 1.25 U AmpliTaq Gold (Applied BioSystem, Branchburg, New Jersey, USA). The PCR conditions were as follows: 94°C, 10 min for 1 cycle; 94°C, 30 s, 60°C, 30 s, 72°C, 30 s for 25 cycles; and finally, 72°C for 7 min.
The samples were then amplified to test for the presence of HPV with different sets of degenerated primers, as described separately for MY09/MY11,38 GP5+/GP6+39 and biotinylated short PCR fragment primer mix located in the L1 region of the HPV genome.40 The PCR conditions for the My09/My11 were as follows: 94°C, 10 min for 1 cycle; 94°C, 30 s, 55°C, 45 s, 72°C, 30 s for 40 cycles, followed by an extension step at 72°C for 7 min. The PCR mix contained 200 μM of each deoxyribonucleotide triphosphate, 40 pmol of each primer, 2 mM MgCl2, 1× PCR buffer and 1.25 U AmpliTaq Gold. For the GP5+/GP6+ primers, the following conditions were used: 94°C, 10 min for 1 cycle; 95°C, 30 s, 44°C, 60 s, 72°C, 90 s for 40 cycles; then a final extension step at 72°C for 7 min. Amplification with short PCR fragment primer mix was carried out as follows: 94°C, 10 min, for 1 cycle; 94°C, 30 s, 52°C, 45 s, 72°C, 45 s for 40 cycles; then a final extension step at 72°C for 7 min. Positive and negative controls were included in each amplification.
HPV typing
HPV typing was carried out using the reverse‐hybridisation assay. The denatured biotinylated amplified product (10 μl) was hybridised with specific oligonucleotide probes, which are immobilised as parallel lines on membrane strips (InnoLiPA, Innogenetics, Ghent, Belgium).40 After hybridisation and stringent washing, streptavidin‐conjugated alkaline phosphatase was added and bound to any biotinylated hybrid previously formed. Incubation with 5‐bromo‐4‐chloro‐3‐indoyl phosphate–nitro blue tetrazolium chromogen yields a purple precipitate that can be visually interpreted. On the basis of the position of the visualised line, it is possible to determine the HPV genotype.40 The following types of HPV were included in the test panel: HPV 6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 70 and 74.
Statistical methods
Statistical analyses were carried out with the SPSS and STATA software packages (SPSS for Windows, V.12.0.1, and STATA/SE V.8.2). Frequency tables for categorical variables were analysed using the χ2 test, the likelihood ratio or Fisher's exact test to assess the significance of the correlation. Bivariate correlations between ordered variables were analysed using Spearman's correlation analysis (Spearman's r). Differences in the means of continuous variables were analysed using non‐parametric tests (Mann–Whitney U) or analysis of variance. Logistic regression models using the stepwise backward approach and likelihood ratio statistics for removal testing were used to analyse the power of different covariates as predictors of the outcome variables (CIN, HR‐HPV), and to calculate crude odds ratios (ORs) and 95% confidence interval (CI). Performance indicators of nm23‐H1 as a marker of CIN or HR‐HPV were calculated with the conventional contingency tables for sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), with 95% CI based on the F‐distribution (±1.96 × standard error). Univariate survival (life‐table) analysis for the outcome measure (clearance or persistence of HPV and overall survival) was based on the Kaplan–Meier method. Multivariate survival analysis was carried out using Cox's proportional hazards model in a backward stepwise manner with the log likelihood ratio significance test, and with the default values for entry and exclusion criteria. The assumption of proportional hazards was checked by log–minus‐log survival plots. In all tests, p<0.05 was considered to be significant.
Results
Table 1 shows the expression of nm23‐H1 related to the grade of the lesion in cone or surgical specimens. An inverse linear relationship was observed between the increasing grade of the lesion and the decreasing intensity of nm23‐H1 (p = 0.001 for linear trend). Reduced expression of nm23‐H1 was associated with CIN3 or cancer at OR 8.72 (95% CI 4.13 to 18.41; p = 0.001), using the two‐tier category of staining (strong or reduced).
Table 1 Expression of nm23‐H1 and grade of cervical lesions.
| Grade of lesion | Expression of nm23‐H1 | |||||
|---|---|---|---|---|---|---|
| Markedly reduced | Slightly reduced | Strong | ||||
| n | % | n | % | n | % | |
| Negative for: CIN | 0 | 0.0 | 1 | 10.0 | 9 | 90.0 |
| CIN1 | 0 | 0.0 | 3 | 15.0 | 17 | 85.0 |
| CIN2 | 2 | 10.0 | 4 | 20.0 | 14 | 70.0 |
| CIN3 | 21 | 21.9 | 37 | 38.5 | 38 | 39.6 |
| SCC | 51 | 38.3 | 48 | 36.1 | 34 | 25.6 |
| Total (n = 279) | 74 | 26.5 | 93 | 33.3 | 112 | 40.1 |
CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma.
Fisher's exact test, p = 0.001 (also for linear trend).
Of the CIN lesions, 70.5% were HR‐HPV positive, in contrast with only 11.1% of those without CIN. HR‐HPV types were even more prevalent in patients with SCC (77.6%), the rest (22.4%) being either negative (4.9%) or those in whom the type of HPV could not be determined (17.5%). Detection of HR‐HPV was associated with SCC at OR 27.25 (95% CI 3.28 to 226.09) and with CIN at OR 19.12 (95% CI 2.31 to 157.81). Among the grades of CIN, the most distinct separation is between CIN2 and CIN3, of which 52.4% and 78.6% are HR‐HPV positive, respectively.
Table 2 shows the expression of nm23‐H1 related to the detection of HR‐HPV in the lesions. Nm23‐H1 staining intensity was only marginally different in HR‐HPV‐positive and HR‐HPV‐negative lesions (p = 0.047), in that the strong expression (equivalent to normal epithelium) was slightly more common in HR‐HPV‐negative cases: 52.1% and 35.5% (OR 1.97, 95% CI 1.14 to 3.42; p = 0.015).
Table 2 Expression of nm23‐H1 related to high‐risk human papillomavirus types in lesions.
| HR‐HPV* | Expression of nm23‐H1 | |||||
|---|---|---|---|---|---|---|
| Markedly reduced | Slightly reduced | Strong | ||||
| n | % | n | % | n | % | |
| Present | 53 | 26.9 | 74 | 37.6 | 70 | 35.5 |
| Absent | 16 | 22.5 | 18 | 25.4 | 37 | 52.1 |
| Total (n = 273) | 69 | 25.7 | 92 | 34.3 | 107 | 39.9 |
HR‐HPV, high‐risk human papillomavirus.
*Also includes cases that were HPV negative; χ2 test, Pearson's p = 0.047.
Table 3 shows the performance indicators calculated for nm23‐H1 as a marker of CIN and HR‐HPV. Reduced nm23‐H1 staining is a 90% specific indicator of CIN, with 98.5% PPV, but strong staining does not rule out CIN, because NPV is only 11.5%. With the CIN3 cut‐off, there is a drop in both specificity and PPV, and a slight gain in sensitivity and NPV. Evidently, nm23‐H1 staining is not a useful marker of HR‐HPV in these lesions.
Table 3 Performance indicators of nm23‐H1 as a marker of cervical intraepithelial neoplasia and high‐risk human papillomavirus type.
| Outcome variable | Performance of nm23‐H1 (95% CI)* | |||
|---|---|---|---|---|
| Sensitivity, % | Specificity, % | PPV, % | NPV, % | |
| CIN† | 48.8 (40.4 to 57.3) | 90.0 (71.4 to 100) | 98.5 (95.6 to 100) | 11.5 (4.4 to 18.6) |
| CIN3‡ | 68.5 (62.5 to 74.6) | 80.0 (68.9 to 91.1) | 94.0 (90.4 to 97.6) | 35.7 (26.8 to 44.5) |
| HR‐HPV | 64.4 (57.7 to 71.1) | 52.1 (40.4 to 63.7) | 78.8 (72.6 to 85.2) | 34.5 (25.6 to 43.5) |
CIN, cervical intraepithelial neoplasia; HR‐HPV, high‐risk human papillomavirus; NPV, negative predictive value; PPV, positive predictive value.
*nm23‐H1 staining (reduced or strong).
†Any grade of CIN (squamous cell carcinoma (SCC) cases excluded).
‡CIN3 cut‐off (SCC cases included).
In all, 41 of 67 (61.2%) of the HPV‐positive women treated for CIN and controlled by serial PCR cleared their HR‐HPV infection by the last PCR assay, with a total of 705 women months at risk and a monthly clearance rate of 5.8% (58/1000 women months at risk). Clearance in 17 of 30 (56.7%) cases with strong nm23‐H1 expression was comparable with that of 22 of 34 (64.7%) cases with reduced staining (p = 0.610). The corresponding figures for virus persistence were 5 of 30 (16.7%) cases and 4 of 34 (11.8%) cases, respectively (p = 0.723). In univariate survival analysis, nm23‐H1 staining was not a significant predictor of either clearance (log rank, p = 0.514) or persistence (p = 0.243) of HR‐HPV types in the cervix after treatment of CIN by large loop excision of the transformation zone.
Of the 150 patients with SCC, 91 (60.7%) were alive and 59 (39.3%) had died during the follow‐up. Nm23‐H1 proved to be a significant predictor of survival in cervical cancer (table 4). The probability of survival was markedly higher (p = 0.022) for patients with strong (normal) nm23‐H1 expression than those with reduced expression (OR 2.82, 95% CI 1.16 to 6.83; p = 0.044). This is also evident in the Kaplan–Meier analysis, where survival was significantly determined by nm23‐H1 expression (p = 0.022; fig 1). HR‐HPV also showed a slightly positive effect on survival in 64.0% of HR‐HPV‐positive women, and 45.2% of HR‐HPV‐negative women were alive (p = 0.062; OR 2.15, 95% CI 0.96 to 4.82). In the Kaplan–Meier analysis, this difference was not significant (p = 0.053). As usual, the International Federation of Gynecology and Obstetrics (FIGO) stage was the most powerful predictor of survival in the Kaplan–Meier analysis (p = 0.001).
Table 4 Expression of nm23‐H1 and prognosis of cervical cancer.
| Expression of nm23‐H1 | Overall survival of patients with cervical cancer | OR (95% CI) | Significance | |||
|---|---|---|---|---|---|---|
| Alive | Dead | |||||
| n | % | n | % | |||
| Strong | 26 | 76.5 | 8 | 23.5 | Reference | |
| Slightly reduced | 24 | 50.0 | 24 | 50.0 | 0.40 (0.15 to 1.06) | p = 0.044 |
| Markedly reduced | 29 | 56.9 | 22 | 43.1 | 0.30 (0.11 to 0.81) | |
| Strong | 26 | 76.5 | 8 | 23.5 | Reference | p = 0.022* |
| Reduced | 53 | 53.5 | 46 | 46.5 | 2.82 (1.16 to 6.83) | |
*Logistic regression.
In multivariate survival (Cox) analysis, nm23‐H1 expression did not prove to be a significant independent prognostic factor. It was removed from the model, when adjusted for age, HR‐HPV, grade of tumour and FIGO stage. In the final Cox model, only the FIGO stage (p = 0.001) and age (p = 0.011) proved to be independent predictors of patient survival. When FIGO stage 1 was used as the reference, the OR for dying from the disease in stage 2 was 5.23 (95% CI 1.78 to 15.36), that in stage 3 was 9.29 (95% CI 3.18 to 27.13) and that in stage 4 was 23.33 (95% CI 6.85 to 79.46). The mean age of women who were alive was 54.2 years as compared with 66.7 years of those who died (p = 0.001).
Discussion
We started a systematic survey of potential biomarkers of HR‐HPV and CIN in our HPV‐Pathogen ISS study.34 Until now, four of the programmed 13 markers of HR‐HPV have been completed: p16INK4,8 ERK1,10 vascular endothelial growth factor (VEGF)‐C11 and survivin.41 Although bearing some interesting links with HR‐HPV and CIN, none of these proved to be an independent prognostic predictor in cervical cancer.8,10,11,41 Recently, a group of proteins, known as the nm23 family, has received considerable attention as a potential anti‐metastatic gene in cervical cancer.12,13,20,21,22,23,24,25 There is no full agreement yet on (a) the relationship between nm23‐H1 expression and grade of CIN, (b) its anti‐metastatic effects or (c) its value as a predictor of survival in cervical cancer.12,13,21,22,23,24,25,27,28,29,30,31,32 Because no previous data are available on the possible interactions between HPV and the nm23 gene family either in vivo or in vitro,1,2,4 we assessed whether nm23‐H1 expression may be of use in predicting any of the several outcome measures: grade of CIN, HR‐HPV type, clearance of the virus and prognosis of cervical cancer, as reported for the other markers.8,10,11,41
Expression of nm23‐H1 as related to grade of CIN has been assessed in a few previous studies.12,13,23,25 Although in some of these studies nm23‐H1 expression was found to increase in parallel with increasing grade of CIN,12,13,25 Lee and Gad23 reported a significant decreasing trend (p = 0.006) from normal epithelium to CIN3 and SCC. This observation is in perfect agreement with the findings of our study, where the reduced expression of nm23‐H1 was shown to increase towards high‐grade CIN, being most frequent among carcinomas (table 1). Indeed, this reduced nm23‐H1 expression was associated with CIN3 or cancer at OR 8.72 (95% CI 4.13 to 18.41; p = 0.001), when compared with CIN1, CIN2 or normal epithelium. The most feasible explanations for these seemingly contradictory observations in the literature could be the (a) different IHC protocols used; (b) different nm23‐H1 antibodies; (c) different systems of grading the IHC staining; and (d) the known problems in grading CIN lesions, particularly the low‐grade CIN.1,2,4
In the light of the available data on the biological functions, it seems feasible to predict that the constitutive expression of this anti‐metastatic protein15,16,17,18,19 should be down regulated in a considerable proportion of high‐grade CIN and invasive cancer as compared with low‐grade CIN and normal epithelium, exactly as reported before23 and also confirmed by our results. In fact, the most dramatic change in nm23‐H1 expression seems to take place on transition from CIN2 to CIN3, when the proportion of normal (strong) expression drops from 70% down to 39% (table 1). These observations are identical with those of Lee and Gad,23 who showed that nm23‐H1 staining was either absent or markedly reduced in all CIN3 lesions, and the difference between CIN1 and CIN3 was significant (p = 0.013). These data suggest that down‐regulated expression of nm23‐H1 is a marker of progressive disease, probably associated with a clonal selection (among the CIN lesions) of cases capable of rapid progression to CIN3 and predestined to develop an invasive phenotype from CIN3.
As it is rarely (if ever) seen in early precancer lesions (CIN1), down‐regulated nm23‐H1 seems to be a late marker of cervical carcinogenesis, and as such markedly differs from those previously analysed in our study.8,10,11,41 In fact, strong or reduced expression of nm23‐H1 distinguished CIN with 90% specificity and 98.5% PPV, but was not particularly sensitive (48.8%; table 3). These figures are slightly inferior to those recently calculated for p16INK4a,8 ERK1,10 survivin41 and VEGF‐C,11 all being 100% specific markers of CIN, with 100% PPV. For nm23‐H1, no comparative data have been previously published. We also tested these “performance indicators” for nm23‐H1 by changing the cut‐off to CIN3, which improved the sensitivity and NPV but slightly compromised the specificity and PPV. In the future, the value of nm23‐H1 as a marker of CIN or cancer will be tested in combination with the other markers, as soon as the analysis of the 13 biomarkers is completed.33,34
In our study, reduced nm23‐H1 staining was associated with HR‐HPV‐positive cases with OR 1.97, 95% CI 1.14 to 3.42 (p = 0.015). This association is much weaker than previously reported for p16INK4a and survivin,8,41 and nm23‐H1 readily dropped out from the multivariate model containing these two independent predictors of HR‐HPV. Accordingly, there seems to be no role for nm23‐H1 as an independent marker of HR‐HPV (table 3), in contrast with p16INK4a, survivin and (to some extent) VEGF‐C, which are all functionally regulated by HR‐HPV oncoproteins.8,11,41 This failure to show any HR‐HPV specificity for nm23‐H1 suggests that this family of proteins is regulated by molecular mechanisms unrelated to oncogenic HPV. At this moment, these regulatory mechanisms remain obscure, however.26,42,43
During the past several years, persistent HR‐HPV infections have received increasing attention as a cause of markedly increased risk of treatment failure of CIN.2,37 Monitoring this risk of disease recurrence after cone treatment with a suitable marker would be of considerable clinical value, and we were interested to see whether nm23‐H1 expression may be of any such predictive value for persistence or clearance of HR‐HPV. In alignment with the failure to find any specificity for HR‐HPV, nm23‐H1 expression did not provide any useful information as a predictor of clearance or persistence of HPV in the cervix after treatment of CIN. With our previous data,8,10,11,41 this implicates that none of the five biomarkers analysed so far (nm23‐H1, VEGF‐C, survivin, ERK1 and p16INK4a) can substitute HPV testing in monitoring the risk of disease recurrence after cone treatment.37
As pointed out earlier, there is no agreement on the value of nm23‐H1 as a predictor of survival in cervical cancer.12,13,21,22,23,24,25,27,28,29,30,31,32 In our study, nm23‐H1 expression was analysed by both univariate (Kaplan–Meier) and multivariate (Cox) survival analysis. Importantly, nm23‐H1 proved to be a major predictor of survival, as 76.5% of the patients with strong nm23‐H1 expression were alive in contrast with 53.5% of those with reduced nm23‐H1 expression (table 4). In Kaplan–Meier analysis, survival was significantly determined by the nm23‐H1 expression with the two‐tier grading (p = 0.022; fig 1). Thus, our results, confirming a positive prognostic value for strong expression of nm23‐H1, are in alignment with the previous studies reporting similar observations in cervical SCC12,13,23,27,29 or adenocarcinoma.28,30 Other studies fail to ascribe any prognostic value for nm23‐H1 expression, however.22,24,28,30,31 In our study, the prognostic value of nm23‐H1 was important only in univariate analysis, but lost its independent value when it was entered into the Cox multivariate model, where the FIGO stage was the single most powerful prognostic predictor. As pointed out, however, it seems most likely that the power of the FIGO stage as an independent prognostic predictor will be hard to exceed by any single marker, and, consequently, any new biomarker showing a predictive value in univariate analysis can be considered to be a potentially useful prognostic predictor in cervical cancer.44
Regarding the mechanisms of the anti‐metastatic effects of nm23‐H1, several possibilities have been implicated.13,27 These known functions of nm23‐H1 include NDP kinase activity, transcription factor, as well as associations with differentiation, proliferation, cell motility and signal transduction.27 It was recently suggested that the nm23 gene instability is probably associated with regulation of tumour progression and would explain the divergent biological role of this protein in different tumours.13 In cervical cancer, nm23‐H1 expression was recently linked with some other markers—that is, H‐ras, cathepsin B (cystein proteinase), collagenases and plasminogen activators.27 Of these, only type IV collagenase (matrix metalloproteinase (MMP)‐2 or gelatinase A) is included in our panel of biomarkers33,34 and was checked for bivariate correlations with nm23‐H1 out of curiosity. Interestingly, a significant inverse correlation was established between nm23‐H1 and MMP‐2 expression (Spearman r −0.167, p = 0.008; data not shown). As recently suggested, interactions between the proteolytic phenotype and expression of nm23‐H1 in cervical cancer seem to exist,27 which would be consonant with this association of down‐regulated nm23‐H1 with up‐regulated MMP‐2 (ie, increased proteolytic activity). More is known about the possible nm23‐H1 pathways in other tumours—for example, in lung cancer, where nm23‐H1 up regulated mRNA and protein levels of β‐catenin, E‐cadherin, TIMP‐1 and CD44s, and down regulated levels of MMP‐2, CD44v6 and VEGF.43 Apart from the above association between nm23‐H1 and MMP‐2, we could readily also confirm the inverse correlation between nm23‐H1 and VEGF‐C (Spearman's r −0.189; p = 0.002) in our series. It remains to be seen whether nm23‐H1 expression is markedly correlated with any of the other biomarkers to be analysed in this series.33,34
Taken together, no evidence was obtained to substantiate direct interactions between HPV and nm23‐H1, the expression of nm23‐H1 being unrelated to HR‐HPV. Nm23‐H1 seems to be a marker of progressive disease, being dramatically down regulated in high‐grade CIN and invasive cancer. No plausible explanation linking this down regulation with HR‐HPV was disclosed, but this down regulation was an adverse prognostic sign in cervical cancer. Whether eventual indirect molecular links to HPV will be disclosed remains to be seen, when the data of our 13 markers are ready for multivariate analysis to predict the intermediate endpoint markers and disease outcome in cervical cancer.
Acknowledgements
This study has been supported by a grant from the Italian Ministry of Health (Ministero della Salute, Ricerca Corrente 1% 2002; Fasc OG/C). The technical and secretarial assistance of Dr Rita Mancini, Mrs Stefania Mochi and Miss Daniela Crupi is gratefully acknowledged.
Abbreviations
CIN - cervical intraepithelial neoplasia
ERK - extracellular signal‐regulated kinase
FIGO - International Federation of Gynecology and Obstetrics
HPV - oncogenic human papillomavirus
HR‐HPV - high‐risk human papillomavirus
IHC - immunohistochemistry
ISS - Istituto Superiore di Sanità
LR‐HPV - low‐risk human papillomavirus
MMP - matrix metalloproteinase
NDP - nucleoside diphosphate
NPV - negative predictive value
PCR - polymerase chain reaction
PPV - positive predictive value
SCC - squamous cell carcinoma
VEGF - vascular endothelial growth factor
Footnotes
Competing interests: None declared.
References
- 1.IARC Monographs on the evaluation of carcinogenic risks to humans. Vol 64. Papillomaviruses. Lyon: IARC, 19951–409. [PMC free article] [PubMed]
- 2.Syrjänen K, Syrjänen S.Papillomavirus infections in human pathology. Chichester: Wiley, 20001–615.
- 3.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 20022342–350. [DOI] [PubMed] [Google Scholar]
- 4.Aubin F, Pretet J L, Mougin C. eds. Papillomavirus humains. Biologie et Pathologie Tumorale. Paris: TEC & DOC, 20031–759.
- 5.Bosch F X, de Sanjose S. Chapter 1: human papillomavirus and cervical cancer—burden and assessment of causality. J Natl Cancer Inst Monogr 2003313–13. [DOI] [PubMed] [Google Scholar]
- 6.Munoz N, Bosch F X, de Sanjose S.et al Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med 2003348518–527. [DOI] [PubMed] [Google Scholar]
- 7.von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer 2002382229–2242. [DOI] [PubMed] [Google Scholar]
- 8.Branca M, Ciotti M, Santini D.et al p16INK4A expression is related to grade of CIN and high‐risk human papillomavirus but does not predict virus clearance after conization or disease outcome. Int J Gynecol Pathol 200423354–365. [DOI] [PubMed] [Google Scholar]
- 9.Giarre M, Caldeira S, Malanchi I.et al Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a‐imposed G1 cell cycle arrest. J Virol 2001754705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branca M, Ciotti M, Santini D.et al Activation of the ERK/MAP kinase pathway in cervical intraepithelial neoplasia is related to grade of the lesion but not to high‐risk human papillomavirus, virus clearance, or prognosis in cervical cancer. Am J Clin Pathol 2004122902–911. [DOI] [PubMed] [Google Scholar]
- 11.Branca M, Giorgi C, Santini D.et al Aberrant expression of vascular endothelial growth factor‐C (VEGF‐C) is related to grade of cervical intraepithelial neoplasia (CIN) and high‐risk human papillomavirus (HPV), but does not predict virus clearance after treatment of CIN or prognosis of cervical cancer. J Clin Pathol 20065940–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P H, Ko J L, Chang H.et al Clinical significance of high nm23‐H1 expression in intraepithelial neoplasia and early‐stage squamous cell carcinoma of the uterine cervix. Gynecol Obstet Invest 20035514–19. [DOI] [PubMed] [Google Scholar]
- 13.Wang P H, Chang H, Ko J L.et al Nm23‐H1 immunohistochemical expression in multisteps of cervical carcinogenesis. Int J Gynecol Cancer 200313325–330. [DOI] [PubMed] [Google Scholar]
- 14.Lombardi D, Lacombe M, Paggi M G. Nm23: unraveling its biology function in cell differentiation. J Cell Physiol 200018244–49. [DOI] [PubMed] [Google Scholar]
- 15.Lacombe M L, Sastre‐Garau X, Lascu I.et al Overexpression of nucleoside diphosphate kinase (Nm23) in solid tumours. Eur J Cancer 1991271302–1307. [DOI] [PubMed] [Google Scholar]
- 16.Zou M, Shi Y, al‐Sedairy S.et al High levels of Nm23 gene expression in advanced stage of thyroid carcinomas. Br J Cancer 199368385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Xu A, Ling X. The expression of nm23‐H1 and p53 protein in nasopharyngeal carcinoma. Lin Chuang Er Bi Yan Hou Ke Za Zhi 199812243–246. [PubMed] [Google Scholar]
- 18.Yang Z, Wen Y, Pu P. The regulation of nm23‐H1/NDPK‐A in different processes of regional lymph node metastases of oral squamous cell carcinomas. Hua Xi Kou Qiang Yi Xue Za Zhi 200321263–266. [PubMed] [Google Scholar]
- 19.Li J, Jin K, Yin S. Study on expression of nm23‐H1 protein/NDPK‐A in laryngeal carcinoma. Lin Chuang Er Bi Yan Hou Ke Za Zhi 199812247–250. [PubMed] [Google Scholar]
- 20.Mandai M, Konishi I, Koshiyama M.et al Altered expression of nm23‐H1 and c‐erbB‐2 proteins have prognostic significance in adenocarcinoma but not in squamous cell carcinoma of the uterine cervix. Cancer 1995752523–2529. [DOI] [PubMed] [Google Scholar]
- 21.Konno R, Sato S, Ito K.et al Immunohistochemical staining of nm23/nucleotide diphosphate kinase in invasive cervical carcinoma tissue and metastatic lymph nodes. Nippon Sanka Fujinka Gakkai Zasshi 199648279–280. [PubMed] [Google Scholar]
- 22.Kristensen G B, Holm R, Abeler V M.et al Evaluation of the prognostic significance of nm23/NDP kinase protein expression in cervical carcinoma: an immunohistochemical study. Gynecol Oncol 199661378–383. [DOI] [PubMed] [Google Scholar]
- 23.Lee C S, Gad J. nm23‐H1 protein immunoreactivity in intraepithelial neoplasia and invasive squamous cell carcinoma of the uterine cervix. Pathol Int 199848806–811. [DOI] [PubMed] [Google Scholar]
- 24.Sarac E, Ayhan A, Ertoy D.et al nm23 expression in carcinoma of the uterine cervix. Eur J Gynaecol Oncol 199819312–315. [PubMed] [Google Scholar]
- 25.Ravazoula P, Aletra C, Kourounis G.et al Immunohistochemical analysis of nm23‐H1 expression in human cervical lesions. Eur J Gynaecol Oncol 200021510–512. [PubMed] [Google Scholar]
- 26.Postel E H, Berberich S J, Flint S J.et al Human c‐myc transcription factor PuF identified as nm23‐H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 1993261478–480. [DOI] [PubMed] [Google Scholar]
- 27.Utrera‐Barillas D, Salcedo‐Vargas M, Gariglio‐Vidal P.et al H‐ras and Nm23‐H1 gene expression and proteolytic activity in squamous cell carcinoma of the uterine cervix. Arch Med Res 200031172–181. [DOI] [PubMed] [Google Scholar]
- 28.Morimura Y, Yanagida K, Hashimoto T.et al Evaluation of immunostaining for MIB1 and nm23 products in uterine cervical adenocarcinoma. Tohoku J Exp Med 1998185185–197. [DOI] [PubMed] [Google Scholar]
- 29.Marone M, Scambia G, Ferrandina G.et al Nm23 expression in endometrial and cervical cancer: inverse correlation with lymph node involvement and myometrial invasion. Br J Cancer 1996741063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Cai S, Yu S. Relationship between nm23‐H1 expression and lymph node metastasis and prognosis in cervical cancer. Zhonghua Fu Chan Ke Za Zhi 199732718–721. [PubMed] [Google Scholar]
- 31.Chen H Y, Hsu C T, Lin W C.et al Prognostic value of nm23 expression in stage IB1 cervical carcinoma. Jpn J Clin Oncol 200131327–332. [DOI] [PubMed] [Google Scholar]
- 32.Busmanis I. Biomarkers in carcinoma of the cervix: emphasis on tissue‐related factors and their potential prognostic factors. Ann Acad Med Singapore 199827671–675. [PubMed] [Google Scholar]
- 33.Syrjänen K J. Immunohistochemistry in assessment of molecular pathogenesis of cervical carcinogenesis. Eur J Gynaecol Oncol 200526118–124. [PubMed] [Google Scholar]
- 34.Branca M, Costa S, Mariani L.et al Assessment of risk factors and human papillomavirus (HPV) related pathogenetic mechanisms of CIN in HIV‐positive and HIV‐negative women. Study design and baseline data of the HPV‐PathogenISS study. Eur J Gynaecol Oncol 200425689–698. [PubMed] [Google Scholar]
- 35.Costa S, Terzano P, Santini D.et al CD44 isoform 6 (CD44v6) is a prognostic indicator of the response to neoadjuvant chemotherapy in cervical carcinoma. Gynecol Oncol 20018067–73. [DOI] [PubMed] [Google Scholar]
- 36.Costa S, Terzano P, Santini D.et al Neoadjuvant chemotherapy in cervical carcinoma: regulators of cell cycle, apoptosis and proliferation as determinants of response to therapy and disease outcome. Am J Clin Pathol 2001116729–737. [DOI] [PubMed] [Google Scholar]
- 37.Costa S, Simone P D, Venturoli S.et al Factors predicting human papillomavirus (HPV) clearance in cervical intraepithelial neoplasia (CIN) lesions treated by conization. Gynecol Oncol 200390358–365. [DOI] [PubMed] [Google Scholar]
- 38.Bauer H M, Greer C E, Manos M. Determination of genital human papillomavirus infection using consensus PCR. In: Herrington CS, McGee JOD, eds. Diagnostic molecular pathology: a practical approach. Oxford: Oxford University Press, 1992132–152.
- 39.Karlsen F, Kalantari M, Jenkins A.et al Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol 1996342095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleter B, van Doorn L J, Scrauwen L.et al Development and clinical evaluation of a highly sensitive PCR‐reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 1999372508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branca M, Giorgi C, Santini D.et al Aberrant expression of a novel inhibitor of apoptosis (Survivin) is related to grade of cervical intraepithelial neoplasia (CIN), but does not predict virus clearance after cone or prognosis in cervical cancer. Am J Clin Pathol 20051241–9. [Google Scholar]
- 42.Shi Y, Parhar R S, Zou M.et al Differential nm23 gene expression at the fetal‐maternal interface. Br J Cancer 199470440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Che G W, Zhou Q H, Zhu W.et al Molecular mechanism of reversing metastatic phenotype in human high‐metastatic large cell lung cancer cell line L9981 by nm23‐H1. Ai Zheng 200524278–284. [PubMed] [Google Scholar]
- 44.Syrjänen K J. Summary statement on prognostic factors in carcinoma of the uterine cervix. CME J Gynec Oncol 20038298–301. [Google Scholar]