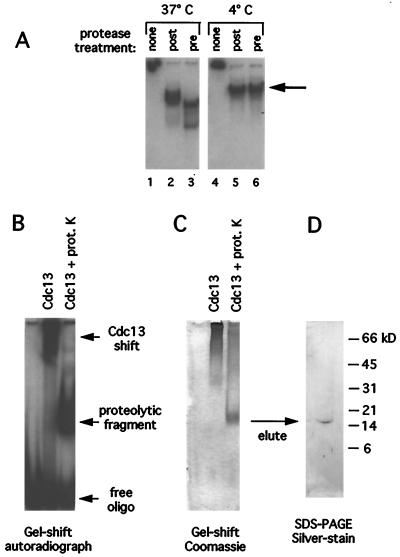
Figure 1.
Proteolysis mapping of the Cdc13 DBD. (A) Telomeric DNA [1 nM end-labeled d(TGTGTGGG)3 oligomer], 5 μg of Cdc13 protein, and proteinase K [added either before (lanes 3 and 6) or after (lanes 2 and 5) addition of oligonucleotide] were assembled on ice, and reactions were incubated at 37°C (lanes 1–3) or 4°C (lanes 4–6) for 15 min. Reaction products were run on 10% polyacrylamide/TBE gels at 4°C; under these conditions, the full length Cdc13p:DNA complex does not enter the gel and is retained in the well (lanes 1 and 4). The large arrow indicates the proteolysis product examined in more detail in Fig. 2. (B) Autoradiograph of a Cdc13p-DNA gel shift [2 μg of Cdc13p protein and 2 μM 5′ end-labeled d(TGTGTGGG)3 oligomer] with or without subsequent proteinase K digestion at 0°C, resolved on a nondenaturing 8% polyacrylamide-TBE gel. prot K, proteinase K. (C) The same gel as in B, fixed and Coomassie-stained. (D) Silver-stained 15% SDS/PAGE tricine gel showing protein eluted from a gel slice containing a proteolytic fragment identical to the one visible in C, with the reaction volume scaled up by a factor of 10.