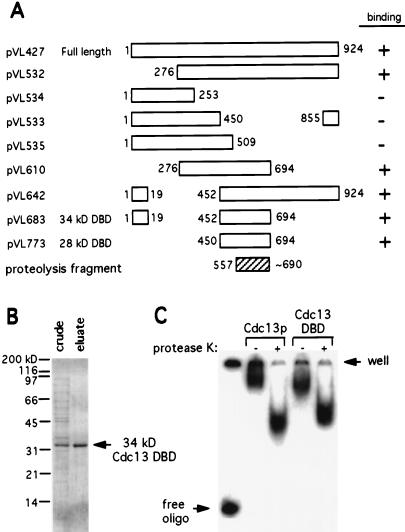
Figure 2.
Deletion mapping and expression of the Cdc13 DBD. (A) DNA binding ability of deletion derivatives of Cdc13, as assessed by DNA gel shift assays with E. coli crude extracts and 5′ end labeled d(TGTGTGGG)3 oligomer; all constructs indicated as “+” exhibited telomeric DNA binding activity comparable to that of the full length protein (data not shown). The N-terminal and predicted C-terminal boundaries of the proteolysis fragment characterized in Fig. 1 (shaded box) are shown for comparison. pVL610 and pVL683 encode additional recombinant material at the C terminus not indicated in the figure (see Materials and Methods for details). (B) Expression of the 34-kDa recombinant His6-tagged Cdc13 DBD (pVL683) in E. coli. Crude extract and eluate samples were resolved on 12% SDS/PAGE and were stained with Coomassie. (C) DNA gel shift reactions, with either the 34-kDa Cdc13 DBD or full-length Cdc13p, with or without proteinase K treatment, were resolved on 8% polyacrylamide/TBE.