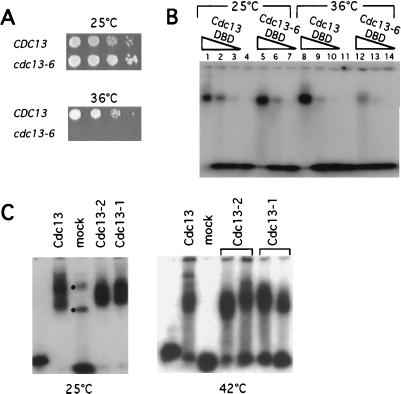
Figure 3.
Identification of a thermolabile DNA binding mutation in Cdc13p. (A) Ten-fold serial dilutions of a CDC13+ yeast strain (cdc13-Δ/pVL440) or a cdc13-6 strain (cdc13-Δ/pVL1295) were spotted onto rich media plates and were grown for 48 h at either 25°C or 36°C. (B) Telomeric DNA gel shift assays with 20 nM (TGTGTGGG)2 and purified Cdc13 DBD (lanes 1–3 and lanes 8–10, at 3 μg, 0.75 μg, and 0.15 μg for each set of three lanes) or Cdc13-6 DBD (lanes 5–7 and lanes 12–14, at 2 μg, 0.5 μg, and 0.1 μg for each set of three lanes); reaction mixtures containing protein and reaction buffer were incubated at 25°C (lanes 1–7) or 37°C (lanes 8–14) for 5 min, followed by addition of end-labeled telomeric substrate and incubation for an additional 15 min at the relevant temperature, and were loaded immediately onto 5% polyacrylamide/TBE gel run at 25°C. Incubation at 37°C did not result in degradation of the Cdc13-6 DBD protein (data not shown). (C) Telomeric DNA gel shift assays with equivalent volumes of partially purified Cdc13, Cdc13-2, Cdc13-1, or mock protein preparations and 1 nM end-labeled yeast d(TGTGTGGG)3 oligomer. Binding reactions and 5% polyacrylamide/TBE gels were either incubated and run at 25°C (Left) or were incubated and run at 42°C, using a prewarmed gel (Right). Dots in the mock protein lane of the 25°C gel correspond to contaminating E. coli DNA binding activity (or activities).