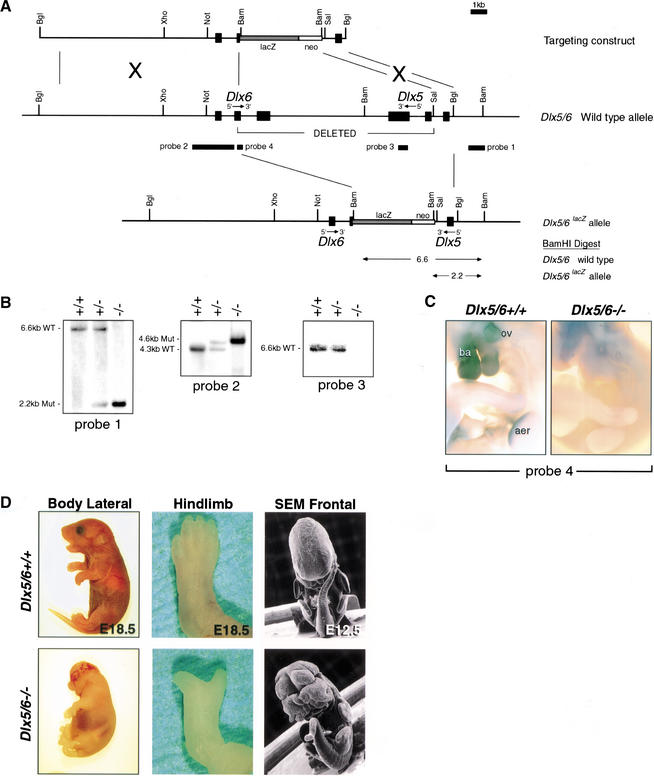
Figure 1.
Simultaneous disruption of Dlx5 and Dlx6 by homologous recombination, genotyping, mRNA, and morphological analyses of embryos carrying a Dlx5/6 null allele. (A) Structure of the genomic Dlx5 and Dlx6 loci, ires.lacZ.neo targeting vector, and mutant allele following homologous recombination. Exons are depicted as black boxes. The transcriptional orientation of each gene is indicated. The probes employed for Southern and RNA in situ analyses are indicated with black rectangles. (B) Southern blot analysis of embryonic genomic DNA isolated from the yolk-sac was digested with BamHI and then hybridized with a 5′ (probe 1) in the undeleted region of Dlx5, 5′ (probe 2) in the undeleted region of Dlx6, or 3′ (probe 3) in the deleted region of Dlx5. The wild-type and mutant alleles were detected as 6.6 kb and 2.2 kb fragments, respectively, with probe 1 and 4.3 kb and 4.6 kb, respectively, with probe 2. The wild-type allele was detected as a 6.6 kb fragment and the mutant allele was undetected with probe 3. (C) Whole-mount in situ hybridization of E10.5 wild-type and Dlx5/6−/− embryos using probe 4. Dlx5/6−/− embryos lack probe 4 expression as probe 4 falls within the region deleted in the Dlx5/6−/− embryos. (D) Gross appearance of wild-type and Dlx5/6−/− embryos and hindlimbs at E18.5. Dlx5/6−/− embryos show an overall reduced size, exencephaly, kinked tail vertebrae, and split hindlimbs. Scanning electron microscopy (SEM) of whole-mount wild-type and Dlx5/6−/− embryos at E12.5, highlighting the craniofacial, tail, and limb malformations already clearly visible at this stage. Abbreviations: aer, apical ectodermal ridge; ba, branchial arches; ov, otic vesicle.