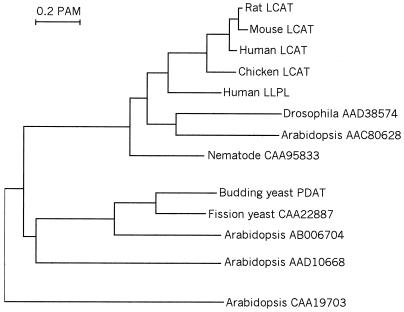
Figure 5.
Evolutionary dendrogram showing LCAT- and PDAT-related proteins from different eukaryotes. The dendrogram was calculated from aligned protein sequences corresponding to amino acid residues 174–335 in yeast PDAT. The clustalx multiple alignment program (28) was used with default settings to align the sequences and compute pairwise alignment scores. An unrooted tree was then obtained from these scores by using the neighbor-joining method (29), with correction for multiple substitutions and exclusion of gapped positions.