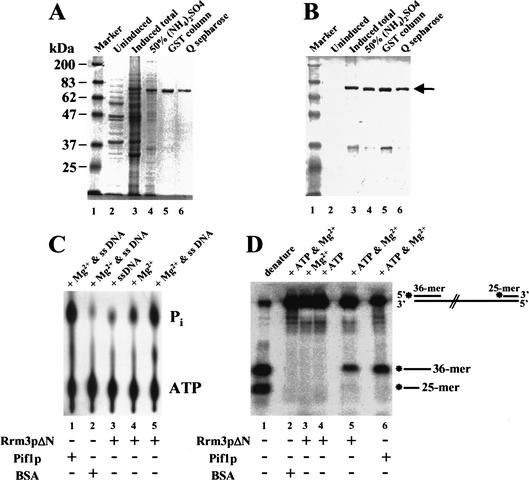
Figure 1.
Recombinant Rrm3 protein has ATPase and 5′ to 3′ DNA helicase activity. A truncated form of Rrm3p (Rrm3pΔN) was fused to GST and expressed in Saccharomyces cerevisiae under control of the galactose-inducible GAL1 promoter. Yeast proteins were resolved by 8% SDS-PAGE and detected by (A) Coomassie blue staining or (B) immunoblotting. In A and B, lane 1 is size markers in kilodaltons. (Because covalent coupling of marker proteins to the blue dye used for visualization affects their mobility in SDS-PAGE, the 83-kD marker protein has a slower mobility than the 87-kD Rrm3pΔN.) Lanes 2 and 3 contain a crude extract of proteins from glucose grown (lane 2) or galactose grown (lane 3) cells. Lanes 4–6 contain proteins from galactose-grown cells after sequential purification by 50% ammonium sulfate precipitation (lane 4), fractionation on Glutathione sepharose 4B (lane 5), and fractionation on Q-sepharose (lane 6). (C) The ATPase reaction products were developed in a polyethylimine (PEI) cellulose plate and visualized on a Molecular Dynamics PhosphorImager. Lane 5 contains [γ-P32]ATP, M13 single-stranded DNA, Mg2+, and Rrm3pΔN. The other lanes were the same except lane 1 had Pif1p in place of Rrm3pΔN; lane 2 had BSA in place of Rrm3pΔN; lane 3 had no Mg2+; and lane 4 had no single-stranded DNA. (D) For the helicase assay, the DNA substrate was linear M13 single-stranded DNA to which kinase labeled 36-mer and 25-mer oligonucleotides had been annealed. Lane 1 contains the heat-denatured DNA substrate; lane 5 contains Rrm3pΔN, the DNA substrate, ATP and Mg2+. The other lanes are the same as lane 5 except lane 2 contains BSA in place of Rrm3pΔN; lane 3 has no ATP; lane 4 has no Mg2+; and lane 6 contains Pif1p in place of Rrm3pΔN.