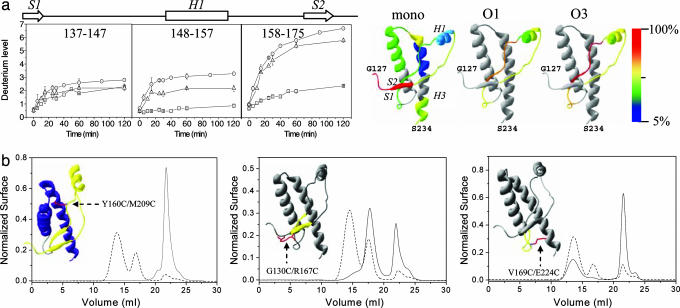
Fig. 4.
Conformational dynamics during PrP oligomerization. (a) H/D exchanges in the OvPrP monomer and oligomers. (Left) Number of deuteriums incorporated for the exchange kinetics, as analyzed by MS, in peptides generated from pepsin digestion of the monomer (squares), O3 (circles), and O1 (triangles) after incubation in a deuterated buffer (from 0 to 120 min). The error bars result from three independent experiments. (Right) Deuterium incorporation after 2 h, normalized to peptide length and averaged between peptides spanning over a common region (see SI Fig. 8 for each individual peptide), is visualized on the OvPrP structure, revealing an increase of accessibility and structural changes near the H1–S2 region. (b) Effect of intramolecular covalent bonds on OvPrP oligomerization. (Left) After 90 min at 50°C, the wild-type OvPrP monomer (80 μM) oligomerizes to yield the O1, O2, and O3 oligomers, which elute as two peaks (dotted line). In contrast, the Y160C/M209C mutant (80 μM) does not generate any oligomeric species in the same conditions (solid line). (Center) In the same conditions, the G130C/R167C mutant (80 μM) oligomerizes to generate major amounts of O3, low amounts of O2 (eluting at 15 ml), and no O1. (Right) The V169C/E224C mutant (100 μM) forms a mixture of O2 and O1 and no O3 oligomer, whereas in the same conditions wild type generates O3 oligomers. Insets show the positions of the additional disulfide bonds.