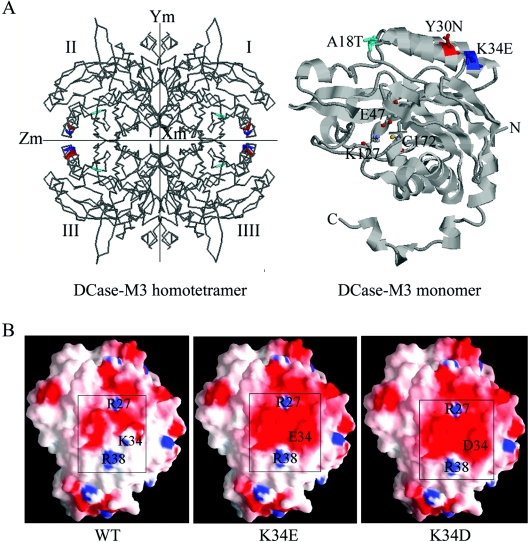
Figure 5. Model of DCase based on its similarity with DCase of Agrobacterium sp. KNK712.
(A) The three-dimensional structure of the DCase-M3 homotetramer and monomer. In the DCase-M3 homotetramer (left-hand panel), the three replacements are coloured: A18T (cyan), Y30N (red) and K34E (blue). In the DCase-M3 monomer (right-hand panel), the three mutations in DCase-M3 are indicated as space-filling models including A18T, Y30N and K34E, which are located on the surface of the protein. The catalytically important residues (Glu47, Lys127 and Cys172) are displayed as ball and stick model, and the N- and C-termini are also labelled in the structure. The Figure was generated using RasMol. (B) Charge distribution of the region near the amino acid at position 34 on the molecular surface of modelled WT and evolved DCases (K34E and K34D). The electrostatic potential is coloured: uncharged amino acids (white), negative amino acids (red) and positive amino acids (blue). All Figures were generated using GRASP.