Abstract
Bloom's syndrome (BS) is an autosomal recessive disorder associated with dwarfism, immunodeficiency, reduced fertility, and elevated levels of many types of cancer. BS cells show marked genomic instability; in particular, hyperrecombination between sister chromatids and homologous chromosomes. This instability is thought to result from defective processing of DNA replication intermediates. The gene mutated in BS, BLM, encodes a member of the RecQ family of DExH box DNA helicases, which also includes the Werner's syndrome gene product. We have investigated the mechanism by which BLM suppresses hyperrecombination. Here, we show that BLM selectively binds Holliday junctions in vitro and acts on recombination intermediates containing a Holliday junction to promote ATP-dependent branch migration. We present a model in which BLM disrupts potentially recombinogenic molecules that arise at sites of stalled replication forks. Our results have implications for the role of BLM as an anti-recombinase in the suppression of tumorigenesis.
Bloom's syndrome (BS) is an autosomal recessive disorder with a wide range of symptoms, including immunodeficiency, short stature, subfertility, and a greatly elevated incidence of a broad spectrum of cancers (1, 2). Cells isolated from BS individuals display a variety of abnormalities, with the hallmark feature being hyperrecombination between sister-chromatids and homologous chromosomes. These recombination events can be visualized cytogenetically as sister-chromatid exchanges and quadriradial chromosomes. BS cells also display more general features of genomic instability, such as a mutator phenotype and an elevated frequency of a variety of chromosomal aberrations (1–3).
The gene mutated in BS, BLM, lies on chromosome 15q in humans (4). Analysis of the BLM cDNA revealed that the predicted amino acid sequence of the BLM protein shows significant similarity to the sequences of DNA and RNA helicases (4). Based on these sequence comparisons, and more latterly on functional studies, BLM has been assigned to the so-called RecQ family of DNA helicases, named after the Escherichia coli recQ gene product (reviewed in refs. 5 and 6). Analysis of recQ mutants has indicated that RecQ protein participates in homologous recombination, but acts as a suppressor of illegitimate recombination (7, 8). Moreover, in combination with RecA and SSB, RecQ can initiate DNA recombination reactions and disrupt recombination intermediates (9). Other RecQ family members include Saccharomyces cerevisiae Sgs1p (10, 11), Schizosaccharomyces pombe Rqh1p (12, 13), and the RECQL, WRN, RECQ4, and RECQ5 proteins from human cells (14–17). In vitro studies have shown that many RecQ family proteins are DNA-dependent ATPases and ATP-dependent DNA helicases that translocate in the 3′–5′ direction (5, 6).
BLM is not the only member of the RecQ family of proteins to be implicated in a human genetic disorder. The WRN gene is mutated in the premature aging condition Werner's syndrome (WS) (16), and the RECQ4 gene is defective in some cases of Rothmund–Thomson syndrome (RTS) (18), a rare disorder associated with skin and skeletal abnormalities (19). WS and RTS are also associated with cancer predisposition, although to a more limited extent than is seen with BS. Moreover, in common with BS, cells derived from WS and RTS individuals show genomic instability (19, 20).
The primary defect leading to hyperrecombination in BS cells is thought to be a delay in the maturation of certain DNA replication intermediates (21, 22). One possible role for BLM would be to prevent DNA structures that arise at blocked or collapsed replication forks from being processed into mature recombinants (5, 6). Here, we have investigated one mechanism by which BLM could suppress hyperrecombination in human cells. We show that BLM possesses the ability to selectively recognize Holliday junctions in vitro. Moreover, we show that BLM efficiently promotes ATP-dependent branch migration of Holliday junctions through greater than 2 kb of DNA.
Materials and Methods
Proteins.
Recombinant human BLM, E. coli RuvA, RuvB, and RecA were purified as described previously (23–26). PcrA was a gift from D. Wigley (University of Oxford, Oxford, U.K.). Terminal deoxynucleotidyltransferase, Klenow polymerase, T4 polynucleotide kinase, and restriction enzymes were obtained from commercial suppliers. Protein concentrations are indicated in moles of monomer.
DNA Substrates.
The synthetic four-way junction (X12) was prepared by annealing four 50-mer oligonucleotides (X12-1, 5′-GACGCTGCCGAATTCTGGCTTGCTAGGACATCTTTGCCCACGTTGACCCG; X12-2, 5′-CGGGTCAACGTGGGCAAAGATGTCCTAGCAATGTAATCGTCTATGACGTC; X12-3, 5′-GACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGC; X12-4, 5′-GCGATAGTCTCTAGACAGCATGTCCTAGCAAGCCAGAATTCGGCAGCGTC) as described (27, 28). X12-1 was 5′-32P-labeled before annealing. Linear single-stranded (ss)DNA and blunt-ended double-stranded (ds)DNA were made from oligonucleotides BL3 (5′-AAAATGAGAAAATTCGACCTATCCTTGCGCAGCTCGAGAAGCTCTTACTTTG) or BL3 annealed to BL4 (5′-CAAAGTAAGAGCTTCTCGAGCTGCGCAAGGATAGGTCGAATTTTCTCATTTT). The linear duplex used in Fig. 3 was prepared by annealing X12-1 with its complement (5′-CGGGTCAACGTGGGCAAAGATGTCCTAGCAAGCCAGAATTCGGCAGCGTC). A 3′-tailed substrate was generated by annealing oligonucleotides X12-1 and 3′-Tail (5′-ATGTCCTAGCAATGTAATCGTCTATGACGTC-3′). pAKE-4 M (4674 bp) was derived from pAKE-7Z (26), and contains a new NheI site created after fill-in by Klenow enzyme of the HindIII site in the polylinker. pAKE-4 M was linearized with PstI and 3′-32P-end-labeled with terminal transferase and [α-32P]ddATP. Gapped duplex pDEA-7Z DNA was prepared as described (26). The 488-bp linear duplex was produced by digestion of pYES2 DNA with PstI and 3′-32P-end labeled using terminal transferase. All DNA concentrations are indicated in moles of substrate.
Figure 3.
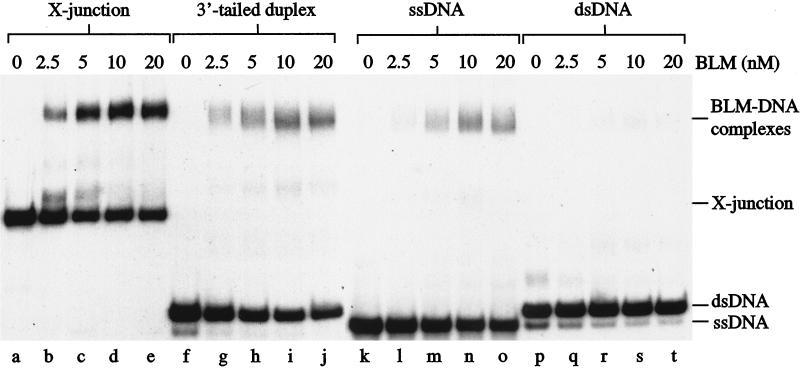
BLM binds selectively to synthetic X-junctions. Band-shift assays were carried out as described in Methods using 0.5 nM substrate representing either 32P-labeled X-junction (lanes a–e), 3′-tailed duplex DNA (lanes f–j), ssDNA (lanes k–o), or blunt-ended dsDNA (lanes p–t), together with the indicated concentrations of BLM.
Branch Migration Assays.
α-Structures were prepared by RecA-mediated strand-exchange, then deproteinized and purified as described (26). Reactions (15 μl) for BLM-mediated branch migration contained 32P-labeled recombination intermediates (0.13 nM) in 20 mM Tris⋅HCl, pH 7.5, 1.25 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, and 1 mM DTT. PcrA reactions were carried out in 20 mM Tris⋅HCl, pH 7.5, 3 mM MgCl2, 100 mM NaCl, 10% glycerol, 2 mM ATP, 0.1 mg/ml BSA, and 5 mM DTT. RuvAB reactions contained RuvA (40 nM) and RuvB (20 nM) in 20 mM Tris⋅HCl, pH 7.5, 15 mM MgCl2, 2 mM ATP, 0.1 mg/ml BSA, and 1 mM DTT. After 45 min at 37°C, reactions were stopped, deproteinized, and 32P-labeled products were visualized by electrophoresis through 1% agarose gels and autoradiography. Gels were run in TAE buffer containing 0.5 μg/ml ethidium bromide (26).
Band-Shift Assays.
Proteins and DNA were incubated in 20 mM triethanolamine⋅HCl, pH 7.5, 2 mM MgCl2, 1 mM adenosine 5′-[γ-thio]triphosphate (ATPγS), 0.1 mg/ml BSA, and 1 mM DTT at room temperature for 20 min. Protein-DNA complexes were fixed with glutaraldehyde (29) and visualized by autoradiography after PAGE through a 5% gel.
X-Junction Dissociation Assays.
Proteins and DNA were incubated in 50 mM Tris⋅HCl, pH 7.5, 2 mM MgCl2, 2 mM ATP, and 0.1 mg/ml BSA. After incubation at 37°C, samples were separated on 10% nondenaturing PAGE gels. Products were visualized by autoradiography.
Results and Discussion
BLM Promotes Branch Migration of Holliday Junctions.
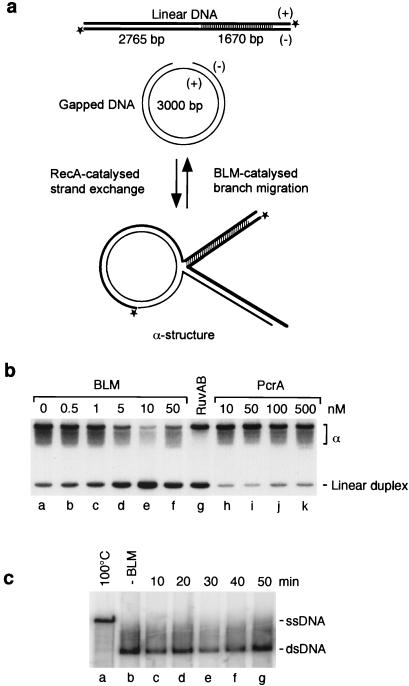
Cells from BS individuals show elevated levels of recombination between sister chromatids and homologous chromosomes, suggesting that BLM is required for the prevention of homologous recombination events (1, 2, 30). For example, BLM might disrupt Holliday junctions, preventing their subsequent processing into recombinant products. To address this possibility, we investigated whether BLM could interact with recombination intermediates and promote in vitro branch migration reactions similar to those mediated by E. coli RuvAB or RecG (31, 32). To do this, α-structures containing Holliday junctions were prepared by RecA-mediated DNA strand exchange between gapped circular and partially homologous 32P-labeled linear duplex DNA (Fig. 1a). Using these substrates, strand exchange proceeds for 2765 bp, but is prevented from reaching completion by the presence of a 1670-bp region of sequence heterology at the distal end of the linear DNA. RuvAB and RecG promote branch migration reactions resulting in the dissociation of the recombination intermediates, regenerating the starting substrates (gapped circular and 32P-labeled linear duplex DNA). The formation of the linear duplex, detected by agarose gel electrophoresis and autoradiography, is the signature of branch migration activity. We found that BLM catalyzed efficient branch migration (Fig. 1b, lanes b–f) in a manner similar to that promoted by RuvAB (lane g). The formation of branch migration products depended on ATP hydrolysis; neither adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) nor adenosine 5′-[γ-thio]triphosphate (ATPγS) could substitute for ATP (Fig. 2 a and b, and data not shown). Control reactions showed that a different DNA helicase (PcrA), which, like BLM, shows a 3′–5′ unwinding polarity, but is also capable of unwinding DNA from blunt ends (23, 33), did not promote branch migration (Fig. 1b, lanes h–k). We confirmed that PcrA was active in standard DNA-unwinding assays (data not shown). Consistent with a specific interaction of BLM with the Holliday junction leading to branch migration over an extended length of DNA (>2.5 kb), BLM was unable to catalyze conventional unwinding of a far smaller (488 bp) 3′-tailed restriction fragment under the same reaction conditions (Fig. 1c). Indeed, in work to be published elsewhere (Brosh et al., unpublished observations), it has been shown that, in the absence of a ssDNA binding protein, BLM is incapable of unwinding 3′ tailed substrates of approximately 250 bp.
Figure 1.
Branch migration of recombination intermediates catalyzed by BLM. (a) Schematic diagram indicating the formation of recombination intermediates (α-structures) by RecA and BLM-mediated branch migration back to the starting substrates. *, 32P labels. (b) Recombination intermediates were incubated with BLM, RuvAB, or PcrA as indicated, and the 32P-labeled products were analyzed as described in Methods. The linear duplex product of branch migration (L) is indicated. (c) Time course showing that BLM (20 nM) is unable to unwind a 488-bp linear duplex (1 nM).
Figure 2.
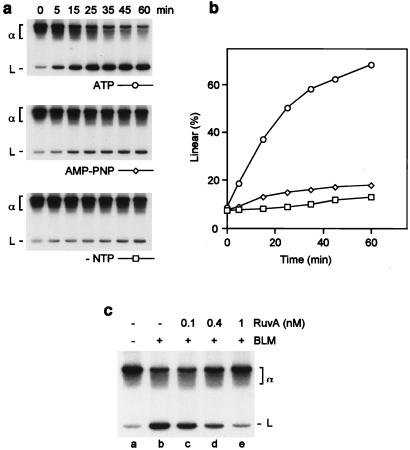
Effect of ATP and RuvA on BLM-mediated branch migration. Reactions were carried out as described in Fig. 1. (a) Time courses were conducted in standard buffer (containing ATP), or in buffer from which ATP was omitted or replaced with 2 mM adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP). (b) The formation of 32P-labeled branch migration products in a was quantified by phosphorimaging and expressed as a percentage of total radiolabel. (c) Recombination intermediates were preincubated with or without RuvA, as indicated, for 3 min on ice. BLM (18 nM) was then added, and reactions were incubated at 37°C for 45 min. Branch migration products were analyzed as in Fig. 1.
Specificity of the BLM–Holliday Junction Interaction.
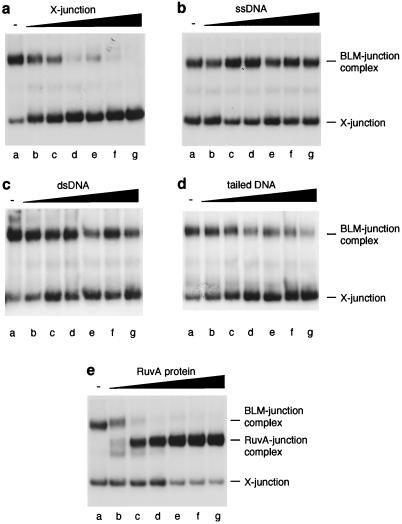
The motor of branch migration in E. coli, RuvB, is relatively inefficient at promoting Holliday junction branch migration, but is stimulated by an association with RuvA, which serves to target RuvB to the junction by virtue of its junction-specific binding activity (28, 31, 34). We found that a short preincubation of the α-structure with RuvA prevented BLM-catalyzed branch migration, indicating that BLM interacts with the Holliday junction to promote branch migration (Fig. 2c, lanes b–e). Hence, we next used band-shift assays to determine whether BLM interacts directly with Holliday junctions. Using 32P-labeled synthetic four-way junctions (X-junctions) that model Holliday junctions, we found that BLM bound X-junction DNA in a protein concentration-dependent manner (Fig. 3, lanes a–e). To assess whether BLM was recognizing the duplex arms of the X-junction, the blunt-ended termini of the arms, or the crossover itself, we analyzed whether BLM could bind under the same band-shift assay conditions to blunt-ended, linear duplex DNA. BLM failed to form a stable complex with linear duplex DNA with a sequence identical to that of one of the “arms” of the X-junction as well as to a similar DNA molecule of unrelated sequence (Fig. 3, lanes p–t; and data not shown), indicating that BLM specifically interacts with the crossover present in the synthetic Holliday junction. Next, we studied the relative efficiency with which BLM binds X-junction DNA, a 3′-tailed duplex (a standard substrate used for analysis of DNA unwinding by 3′–5′ DNA helicases such as BLM) and ssDNA. BLM formed a specific complex with each of these DNA molecules (Fig. 3, lanes a–e, f–j, and k–o, respectively). On the basis of averaging the results of three independent experiments, the level of complex formation at an equivalent BLM concentration was approximately 2- and 5-fold higher with the X-junction DNA than with 3′-tailed DNA and ssDNA, respectively.
That BLM interacts selectively with Holliday junctions was supported by observations showing that unlabeled X-junctions (Fig. 4a, lanes b–g), but not ssDNA (Fig. 4b, lanes b–g), dsDNA (Fig. 4c, lanes b–g), or 3′-tailed duplex DNA (Fig. 4d, lanes b–g) acted as an efficient competitor for the binding of BLM to X-junctions. On the basis of averaging the results of three to five independent experiments in each case, the concentration of competitor required to reduce binding of BLM to the X-junction DNA to 50% of control values was as follows: X-junction, 7.6 μM; ssDNA, >100 μM; dsDNA, >100 μM; 3′-tailed DNA, 46.8 μM. Hence, based on these analyses, it would appear that BLM shows an affinity for X-junction DNA that is >15-fold higher than for ssDNA or dsDNA, and approximately 6-fold higher than for 3′-tailed DNA. Consistent with the notion that BLM recognizes the DNA crossover, the RuvA protein was found to compete efficiently with BLM for binding to the X-junction (Fig. 4e, lanes b–g; <3 μM RuvA required to reduce BLM binding by 50%). Analysis of this competition indicated that the affinity of BLM for the junction was less than that observed with RuvA, because, in the presence of a 5-fold excess of BLM over RuvA (based on moles of monomer), RuvA-junction complexes predominated (Fig. 4e, lane d).
Figure 4.
Competition for binding of BLM to the X-junction. Band-shift assays were carried out using BLM (68 nM) and 32P-labeled X-junction (0.5 nM) as described in Methods. (a) Unlabeled X-junction competitor; (b) ssDNA competitor; (c) dsDNA competitor; (d) 3′-tailed duplex competitor; (e) RuvA competitor. Lanes: a, no competitor; b, 3.1 nM; c, 6.3 nM; d, 12.5 nM; e, 25 nM; f, 50 nM; g, 100 nM competitor in each case.
In addition to dissociating recombination intermediates made by RecA (Figs. 1 and 2), we also observed that BLM could promote ATP-dependent dissociation of 32P-labeled synthetic X-junctions in vitro. BLM-mediated junction dissociation was found to be blocked by the presence of unlabeled X-junction competitor, but not by 3′-tailed DNA, ssDNA, or dsDNA (data not shown). Moreover, the RuvA protein also efficiently blocked junction dissociation. These data support the notion that BLM exhibits some specificity for Holliday junctions in duplex DNA.
Interestingly, recent studies have shown that BLM (24), like the RuvB branch migration motor (35), forms oligomeric ring structures, despite the absence of extensive primary sequence similarity between the proteins. Further work will be required to reveal whether BLM and RuvB recognize Holliday junctions and catalyze branch migration in a mechanistically similar manner.
A Model for the Role of BLM as a Suppressor of Recombination.
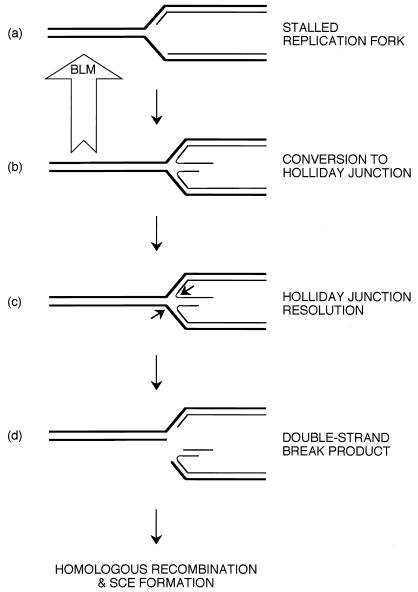
Several lines of evidence indicate that BLM and other members of the RecQ helicase family act during DNA replication; in particular, BS cells show retarded S-phase progression and an accumulation of abnormal replication intermediates (21, 22). Moreover, the high levels of sister-chromatid exchanges that are the hallmark of BS cells occur primarily as a result of replication proceeding on a template that has incorporated bromodeoxyuridine, which is used to reveal the exchanges cytogenetically (36). These data, combined with evidence that BLM serves to prevent “promiscuous” homologous recombination, lead us to propose the model presented in Fig. 5. In this model, BLM acts at sites of stalled or disrupted replication forks to prevent inadvertent or promiscuous recombination. The free DNA ends of the newly synthesized daughter strands at a stalled replication fork may anneal to each other, forming a Holliday junction, as suggested previously from studies of E. coli ruvABC mutants (37). We propose that BLM recognizes the Holliday junction and catalyzes reverse branch migration, leading to the restoration of the replication fork. In this way, the induction of dsDNA breaks, through the action of an as yet unidentified Holliday junction resolvase (equivalent to E. coli RuvC) (38), is minimized. On removal of the blockade, replication can recommence. In the absence of BLM, in BS cells, Holliday junctions are likely to persist during S-phase at sites of stalled forks, allowing junction-resolution to occur, leading to the initiation of double-strand break repair by homologous recombination. Because RecQ and Sgs1 can disrupt synthetic 3- and 4-way junctions in vitro (9, 39), it is possible that the promotion of Holliday junction branch migration is a universal property of RecQ family helicases. Further work will be required to confirm this.
Figure 5.
Model for the role of BLM as an anti-recombinase at sites of blocked replication forks. At sites of stalled forks (a), nascent DNA strands can dissociate from the template and anneal to each other, generating a Holliday junction (b). If this structure is not destroyed by the reverse branch migration activity of BLM, as indicated, junction resolution can occur (c) with the generation of a dsDNA break (d) and the subsequent formation of recombinants. See text for details.
A question that arises from this model is how directionality might be imposed on the branch migration reaction, because there would be a requirement for branch migration to proceed in a reverse direction in order for the junction to be dissociated, as has been shown previously for RecG (32). Forward branch migration would cause the junction to translocate away from the point where replication fork progression was halted. For BLM to act in a directional manner would likely require the participation of other factors to effect the asymmetric binding of BLM to the junction. For example, proteins that are already associated with the fork might impose a bias in how BLM binds junctions. It may be significant in this regard that functional interactions have been identified between BLM and replication protein A, the ssDNA binding protein (Brosh et al., unpublished observations).
Our model has similarities to and differences from the model proposed recently by Courcelle and Hanawalt (40). In their model, based on an analysis of E. coli recQ mutants, it was proposed that the RecQ helicase in conjunction with RecJ nuclease selectively degrade the nascent lagging strand at blocked replication forks both to prevent illegitimate recombination and to stabilize the fork to ensure that accurate recovery of replication can occur when the blockade has been removed. In our model for the action of BLM in human cells, BLM achieves a similar goal—that of preventing “promiscuous” recombination events and stabilization of the fork—but by the route of eliminating a Holliday junction recombination intermediate. Given that there are at least five members of the RecQ family in human cells, it is conceivable that a family member other than BLM is involved in a degradative reaction analogous to that proposed for RecQ/RecJ. It may be significant in this regard that the WRN protein is a combined helicase/nuclease (41, 42).
It should be noted that, whereas BLM promotes efficient disruption of recombination intermediates, it clearly is not limited to this role. Previous work has shown that BLM is a general helicase with preference for G-quadruplex DNA (43). Hence, BLM may play at least two roles in the cell: one concerned with Holliday junction interactions, and the other concerned with disruption of DNA secondary structures that impede the progress of protein complexes involved in replication and/or recombination.
In summary, we have shown that the BS gene product can promote branch migration. We suggest that the anti-recombinase role of BLM is important to maintain genome stability and is critical in suppressing tumorigenesis in humans. In future studies, it will be interesting to determine whether WRN and RECQ4, the RecQ family helicases defective in Werner's and Rothmund–Thomson syndromes (16, 18), display similar anti-recombinase activities.
Acknowledgments
We thank our colleagues for helpful discussions and Dale Wigley for PcrA protein. The Imperial Cancer Research Fund and the Human Frontiers Science Program supported this work. J.K.K. is a Boehringer Ingelheim Fonds scholar. A.C. was supported by a European Science Exchange Fellowship from the Royal Society and the Swiss National Foundation.
Abbreviations
- BS
Bloom's syndrome
- ds
double-stranded
- ss
single-stranded
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100448097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100448097
References
- 1.German J. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 2.German J. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 3.German J, Crippa L P, Bloom D. Chromosoma. 1974;48:361–366. doi: 10.1007/BF00290993. [DOI] [PubMed] [Google Scholar]
- 4.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Chakraverty R K, Hickson I D. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Karow J K, Wu L, Hickson I D. Curr Opin Genet Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 8.Hanada K, Utika T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmon F G, Kowalczykowski S C. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt P M, Louis E J, Borts R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 12.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray J M, Lindsay H D, Munday C A, Carr A M. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K, et al. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 16.Yu C, Oshima J, Fu Y, Wijsman E M, Hisama F, Alisch R, Matthews S, Najura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 17.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 18.Kitao S, Shimamoto A, Goto M, Miller R W, Smithson W A, Lindor N M, Furuichi Y. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 19.Vennos E M, James W D. Dermatol Clin. 1995;13:143–150. [PubMed] [Google Scholar]
- 20.Epstein C J, Martin G M, Schultz A L, Motulsky A G. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Lonn U, Lonn S, Nylen U, Winblad G, German J. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 22.Ockey C H, Saffhill R. Carcinogenesis. 1986;7:53–57. doi: 10.1093/carcin/7.1.53. [DOI] [PubMed] [Google Scholar]
- 23.Karow J K, Chakraverty R K, Hickson I D. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 24.Karow J K, Newman R H, Freemont P S, Hickson I D. Curr Biol. 1999;9:597–600. doi: 10.1016/s0960-9822(99)80264-4. [DOI] [PubMed] [Google Scholar]
- 25.Tsaneva I R, Illing G, Lloyd R G, West S C. Mol Gen Genet. 1992;235:1–10. doi: 10.1007/BF00286175. [DOI] [PubMed] [Google Scholar]
- 26.Eggleston A K, Mitchell A H, West S C. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd R G, Sharples G J. Nucleic Acids Res. 1993;21:1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons C A, Tsaneva I, Lloyd R G, West S C. Proc Natl Acad Sci USA. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons C A, West S C. J Mol Biol. 1993;232:397–405. doi: 10.1006/jmbi.1993.1399. [DOI] [PubMed] [Google Scholar]
- 30.Chaganti R S K, Schonberg S, German J. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsaneva I R, Muller B, West S C. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 32.Whitby M C, Ryder L, Lloyd R G. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- 33.Bird L E, Brannigan J A, Subramanya H S, Wigley D B. Nucleic Acids Res. 1998;26:2686–2693. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons C A, Stasiak A, Bennett R J, West S C. Nature (London) 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 35.Stasiak A, Tsaneva I R, West S C, Benson C J, Yu X, Egelman E H. Proc Natl Acad Sci USA. 1994;91:7618–7622. doi: 10.1073/pnas.91.16.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiraishi Y, Yosida T H, Sandberg A A. Proc Natl Acad Sci USA. 1983;80:4369–4373. doi: 10.1073/pnas.80.14.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 38.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 39.Bennett R J, Keck J L, Wang J C. J Mol Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 40.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 41.Huang S, Li B, Gray M D, Oshima J, Mian I S, Campisi J. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen J-C, Gray M D, Oshima J, Kamath-Loeb A S, Fry M, Loeb L A. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 43.Sun H, Karow J K, Hickson I D, Maizels N. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]