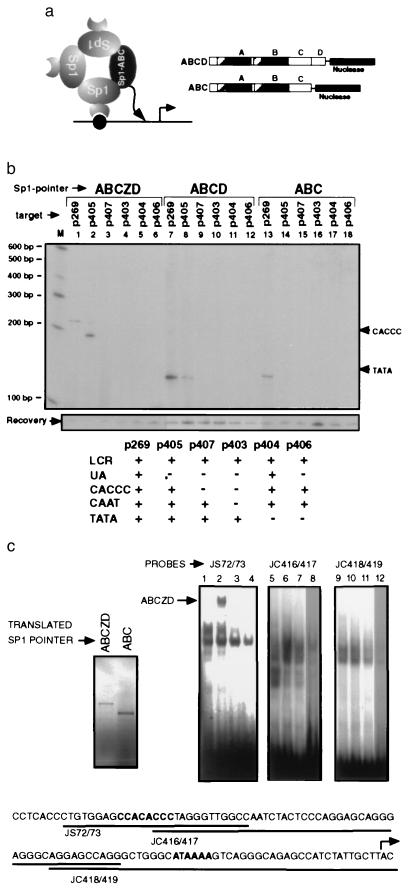
Figure 3.
Promoter elements participate in the formation of a multimeric Sp1 complex. (a) A diagram of hypothetical Sp1–Sp1 interactions that may recruit the Sp1 pointer (solid oval) lacking the DNA binding domain (ABC) is shown on the left and the structure of the Sp1 pointers ABCD and ABC is shown on the right. (b) Recruitment of Sp1 pointers ABCZD (lanes 1–6), ABCD (lanes 7–12), and ABC (lanes 13–18) to target plasmids containing deletions of different promoter elements was analyzed with PIN*POINT. Primer extension on the noncoding strand is shown. The promoter elements that are present (+) and lacking (−) in each of the target plasmids are summarized in the table below. Positions of the distal CACCC box and TATA box are shown on the right. (c) EMSA analysis was performed with in vitro-translated Sp1 pointers ABCZD and ABC (translated product shown in upper left corner) and oligonucleotide probes (JS72/73, JC416/417, and JC418/419) shown below. The proximal CACCC box and TATA box are in boldface type and the transcription initiation site is marked with a bent arrow. EMSA analysis was done with the probes indicated above and rabbit reticulocyte lysate alone (lanes 1, 5, and 9), in vitro-translated Sp1 pointer ABCZD without (lanes 2, 6, and 10) or with competing Sp1 binding oligonucleotide (lanes 3, 7, and 11), and in vitro-translated Sp1 pointer ABC (lanes 4, 8, and 12). The DNA bound ABCZD complex is indicated with an arrow. Competition with a non-Sp1-binding oligonucleotide did not affect this complex, suggesting that it is a sequence specific complex (data not shown).