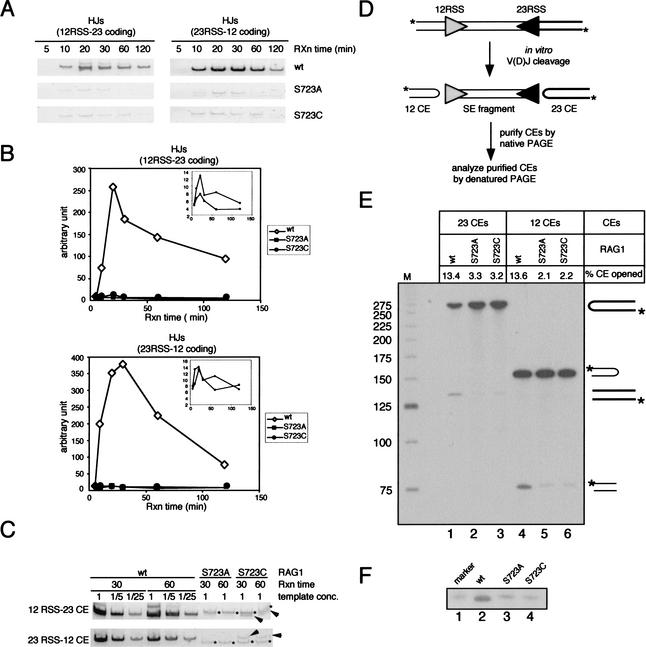
Figure 5.
Postcleavage activities of the RAG proteins in vitro. (A) Hybrid joint formation. Products of the coupled cleavage reactions described in Fig. 1B were used as templates for PCR reactions to amplify hybrid joints. PCR products were resolved on 6% polyacrylamide gels and visualized by staining with cybergreen I. (B) Quantitation of hybrid joints in (A) by FluorImager (Molecular Dynamics). (C) Two time points (30 and 60 min) of the coupled cleavage reaction products were further analyzed by a semiquantitative PCR assay. The reactions containing the wild-type RAG1 protein were subjected to successive fivefold dilutions prior to PCR amplification, whereas the reactions containing the mutant RAG1 proteins were used as templates for PCR without dilution. Asterisks indicate nonspecific PCR products, and arrows indicate specific HJ products. (D) Schematic diagram showing the assay to detect hairpin opening by the RAG proteins in the context of coupled cleavage. Large-scale coupled cleavage reactions were carried out using a 5′ end-labeled PCR substrate, and the resulting coding ends were purified from a 4% native polyacrylamide gel. Purified coding ends were then analyzed on a 10% denaturing gel containing 40% (v/v) formamide, shown in (E). Unprocessed coding ends give rise to bands at 272 nt and 154 nt, whereas the processed coding ends yield products of ∼136 nt and 77 nt. The hairpin opening activity (% CE opened) is indicated above each lane and was determined by calculating the ratio of the processed coding end to the total coding end and multiplying by 100. (F) Purified 12-coding ends were further analyzed by comparing to a synthetic oligonucleotide identical in length and sequence to the top strand of the 12-coding end that has been nicked at the hairpin tip (lane 1). The processed coding ends in the wild-type RAG1 reaction migrated slightly faster than the marker and the processed coding ends from the reactions containing mutant RAG1 proteins (cf. lane 2 and lanes 1,3,4).