Figure 1.
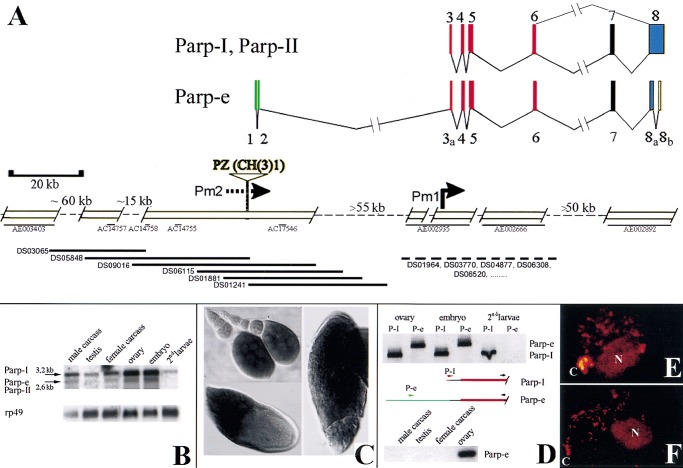
Structure and expression of the Drosophila Parp locus. (A) Deduced genomic structure of the 300-kb Parp region; open boxes are sequenced. The arrangement of the exons encoding Parp-I is shown at top (Uchida et al. 1993; Hanai et al. 1998). (Bottom) The positions of three unlinked Drosophila genomic contigs (thin black lines: AE002935, AE002666, and AE002892) homologous to Parp-I exons are shown at right (Adams et al. 2000). Pm1 indicates the Parp-I promoter deduced from 5′ cDNA sequences (Hanai et al. 1998). A single cDNA isolated from early ovarian stages, GM10715, defines the alternatively spliced Parp-e transcript. The 5′-most 273 bp of GM10715 matches the genomic sequence flanking a P element insertion, CH(3)1 (Zhang and Spradling 1994). A map of this region (left portion of figure) was constructed by chromosome walking using a P1 genomic library (Kimmerly et al. 1996) (bottom, thick black lines). The two resulting scaffolds were sequenced and found to span four small preexisting genomic sequence contigs (thin black lines) and to link to a fifth (AE003403). The color code indicates which portion of PARP is encoded: DNA binding (red), automodification (purple), catalytic (blue), noncoding (green and yellow). (B) Multiple Parp transcripts. A Northern blot of poly(A)-containing RNA from the indicated developmental stages reveals both a 3.2-kb RNA, the size predicted for Parp-I, and a 2.6-kb RNA, the approximate size expected for Parp-II and Parp-e. Parp-homologous RNAs are abundant in both ovaries and embryos, and are reduced but still detectable in second instar larvae and adults. (C) Whole mount in situ hybridization using a 1.4-kb cDNA probe from the DNA-binding domain common to all isoforms labels Parp RNA in nurse cells and in oocytes from stage 9–14 follicles. (D) RT-PCR using isoform-specific primers (see diagrams) that distinguish between Parp-I (or Parp-II) and Parp-e demonstrate that Parp-e is produced in ovaries and embryos, but not at detectable levels in second instar larvae, or in adults outside the ovary. (E) Nuclei are shown from brains of third instar larvae expressing a Parp-I-DsRed fusion gene (see Materials and Methods). Protein is abundant in the chromocenter (C), the nucleolus (N), and at sites within euchromatin. (F) Third instar larval brain nuclei stained with anti-poly(ADP-ribose) antibody 10H show that protein-associated ADP-ribose is found in the same regions as PARP-DsRed.