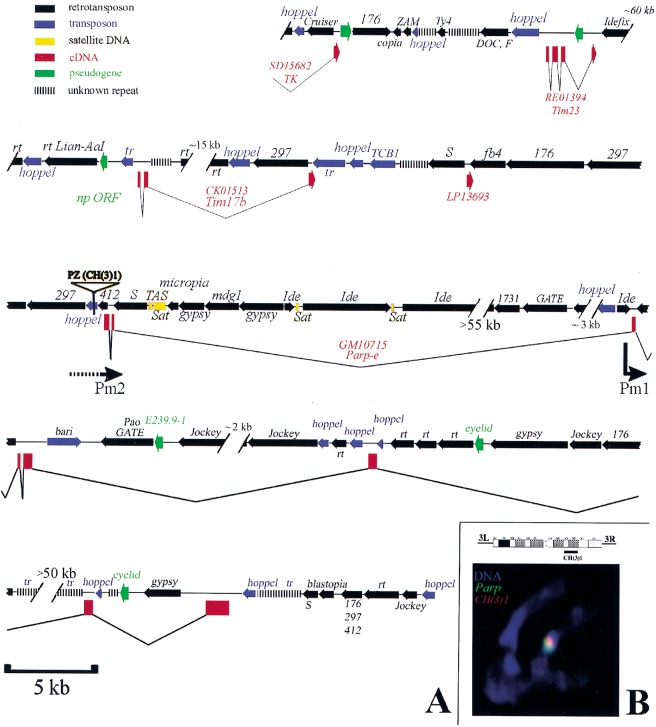
Figure 2.
DNA sequence of the heterochromatic region containing Parp. (A) A diagram summarizing the sequence organization of the region as determined from this study (see Materials and Methods) and from Adams et al. (2000) is shown. Genes defined by cDNAs sequenced as part of this study are shown in red (boxes are exons). The names of retrotransposons (black) and of transposons (blue) are given above the region of homology represented by an arrow (arrowhead: 3′ end). Regions containing only small sequence blocks related to a particular transposon are indicated by parallel bars. The position of the CH(3)1 insertion and the location of the putative Parp promoters Pm1 and Pm2 are indicated. Gaps in the sequence of known or estimated size are represented by hash marks. (B) An ideogram of chromosome 3 heterochromatin shows the cytological region of CH(3)1 insertion (Zhang and Spradling 1994). (Bottom) A chromosome set from a CH(3)1/TM3 third instar larval neuroblast is shown that has been hybridized in situ with Parp cDNA (green) and transposon-specific sequences (red). The partial overlap of the Parp and CH(3)1 sequences indicates that Parp and CH(3)1 are located near each other in 3R heterochromatin. (Note: the TOTO-3 used for this confocal micrograph does not reveal full morphological detail, but chromosomes were also scored using DAPI; CH(3)1 was localized previously to h55 (Zhang and Spradling 1994).