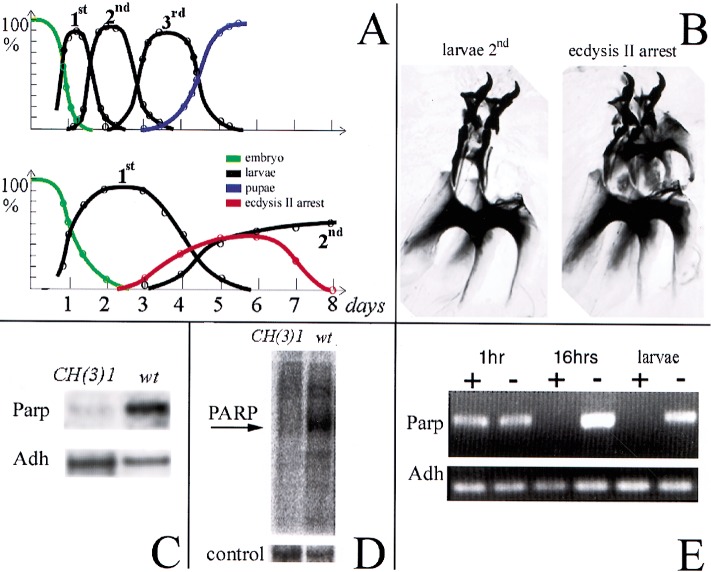
Figure 3.
The CH(3)1 complementation group disrupts Parp expression and activity. (A) Timelines of development of wild-type (top) and CH(3)1 homozygotes (bottom) are shown. The fraction of animals at each developmental stage are plotted as a function of time, revealing the strong developmental delay caused by CH(3)1. (B) Preparation of larval mouth hooks, which distinguish larval instars, are illustrated showing the characteristic appearance of the normal l2 mouthhooks (left) and of mouthhooks from CH(3)1 mutants arrested at the onset of ecdysis 2 (right). (C) Northern blot of poly(A)-containing RNA from wild-type larvae and 4-d-old CH(3)1 larvae showing reduced levels of Parp 3.2-kb mRNA. (D) Proteins labeled by ADP-ribosylation in wild-type (wt) and CH(3)1 mutant larvae. An autoradiogram of a gel of 32P-labeled protein is shown (see Materials and Methods). The prominent band at 117 kD in wild-type has the expected molecular weight of Parp itself. Stained protein in a segment of the same gel is shown as a loading control. (E) RNAi treatment of embryos eliminates detectable Parp mRNA in 16-h embryos and larvae. An RT-PCR assay recognizing all forms of the Parp transcripts is shown; primers specific for the alcohol dehydrogenase gene (Adh) gene serve as a loading control.