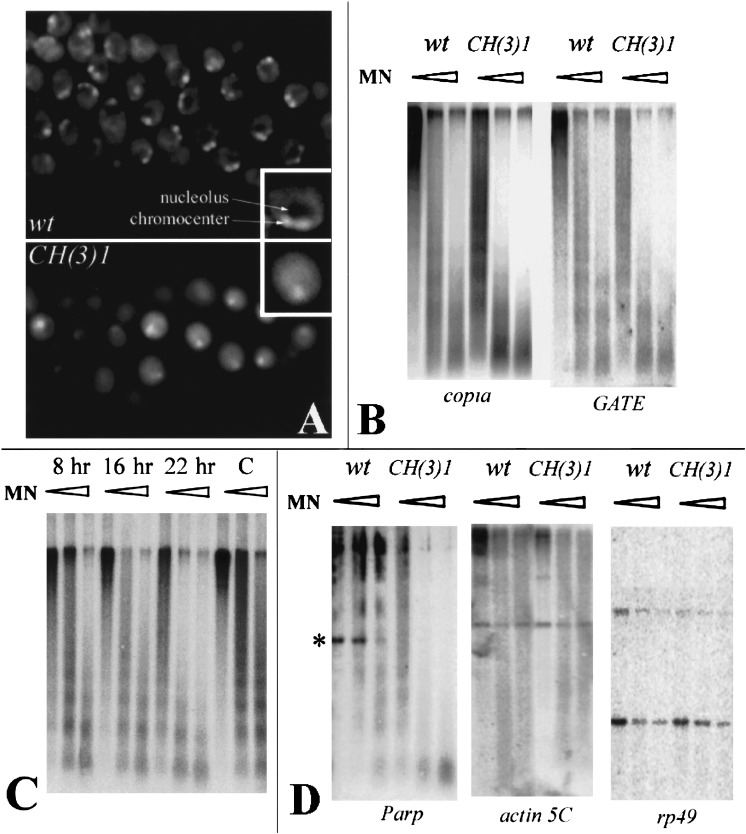
Figure 5.
Parp mutations alter nuclear morphology and chromatin accessibility to nuclease. (A) DAPI-stained nuclei from second instar larval salivary glands of wild-type (top) or CH(3)1 mutants (bottom). A single nucleus is presented at higher magnification in the insets. Nuclei in the mutant appear more diffuse, have a less distinct chromocenter, and lack the region of low DNA density caused by the presence of a normal nucleolus. (B) Nuclei from CH(3)1 mutant larvae were treated with increasing concentrations of micrococcal nuclease (triangles) prior to DNA extraction, digestion with PstI, and analysis on Southern blots probed with a copia or GATE probe. Pst digestion produces no small internal fragment of copia or GATE resolvable within the molecular weight range of the gel. At all concentrations, retrotransposon-specific sequences were far more sensitive to digestion in the mutant. (C) The same analysis as in B was carried out using nuclei at the indicated times after injection of Parp-specific RNAi. Copia sequences from RNAi-injected animals become increasingly sensitive to micrococcal nuclease digestion with increasing time after RNAi injection, compared to buffer-injected controls (C). (D) Micrococcal nuclease assays were carried out as in B and analyzed with a probe from the Parp gene region encoding exons 3, 4, and 5, and with probes specific for the single-copy euchromatic genes actin 5C and rp49. Parp sequences are much more accessible to digestion in the mutant, including a band containing exon 3 and Pm1 (asterisk). To ensure that experiments with heterochromatic and single-copy probes were comparable, the same blot was used for copia, GATE, actin 5C, and rp49. The blot assayed with Parp in D was reprobed with copia as a control and showed the same differential digestion as in B.