Figure 5.
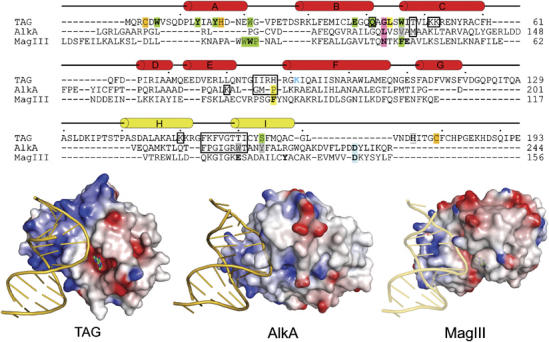
Comparison of 3-methyladenine DNA glycosylases. Top: structure-based sequence alignment of TAG, AlkA, and MagIII shows the relative positions of residues important for DNA binding and base excision. TAG secondary structure elements are shown schematically, with the HhH motif colored yellow. Residues contacting the DNA backbone are boxed, intercalating plug (pink) and wedge (yellow) residues are highlighted, and side chains contacting the estranged base are labeled blue. Side chains confirmed (green) or postulated (gray) to contact 3mA in the base binding pocket are highlighted. Residues verified biochemically to affect substrate binding or catalysis are shown in boldface and the catalytic aspartates in AlkA and MagIII are shaded blue. TAG residues that coordinate Zn2+ are shaded orange. Bottom: crystal structures of TAG/DNA/3mA, AlkA/DNA, and MagIII/3mA (with modeled DNA) are shown. Protein solvent-accessible surfaces are colored according to the electrostatic potential (blue, positive; red, negative). An alternate version of this figure showing all HhH glycosylase/DNA complexes is available as Supplementary data.
