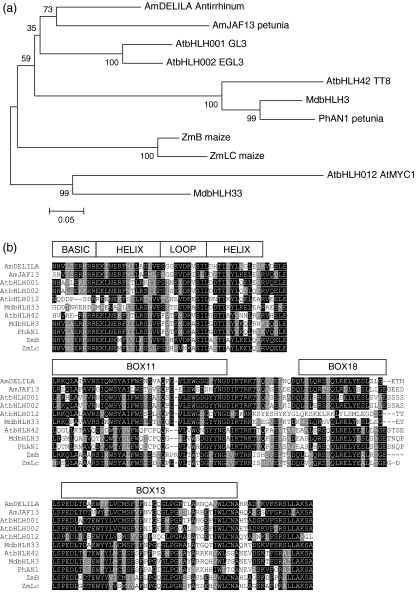
Figure 4.
MdbHLH3 and MdbHLH33 share homology at the bHLH motif with anthocyanin bHLH regulators from other species.
(a) Unrooted phylogenic tree showing that MdbHLH3 clusters in the same group as AtTT8 whilst MdbHLH33 clusters in the same group as AtMYC1. Full-length sequences were aligned using Clustal W (opening = 15, extension = 0.3) in Vector NTI 9.0. Conserved motifs were extracted and re-aligned as above. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004) using a minimum evolution phylogeny test and 1000 bootstrap replicates.
(b) Protein sequence alignment, with the structure of the bHLH binding domain indicated. The accession numbers of these proteins, or translated products, in the GenBank database are as follows: AmDELILA, AAA32663; ATMYC1, BAA11933; EGL1, Q9CAD0; G13, NP680372; PhJAF13, AAC39455; MdbHLH3, CN934367; MdbHLH33, DQ266451; PhAN1, AAG25928; AtTT8, CAC14865; ZmB, CAA40544; ZmLC, AAA33504.
(c) N-terminus of the same bHLH subset showing regions (boxes 11, 18 and 13) conserved within the bHLH IIIf clade (according to Heim et al., 2003). Identical residues are shown in black, conserved residues in dark grey, and similar residues in light grey.