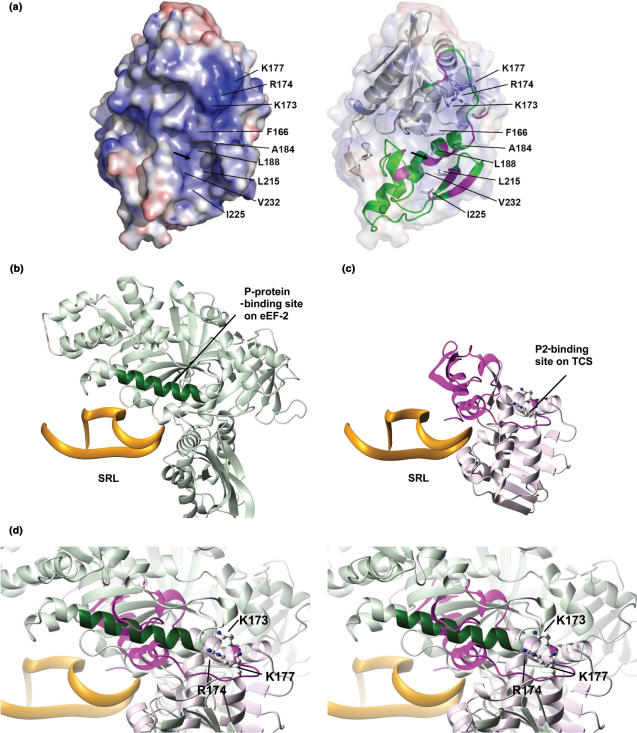
Figure 7.
Comparison of P-protein-binding sites on eEF2 and TCS. (a) Putative P2-binding surface of TCS. The electrostatic calculation was performed using the program APBS (44) and visualized by PyMOL (45), where positive and negative potential surface is colour-coded blue and red, respectively, at ±10 kT. K173, R174 and K177 contribute to a strongly positive-charged surface which may interact with the DDD motif of P2. The arrow indicates the location of the hydrophobic pocket constituted by F166, A184, L188, L215, I225 and V232. A ribbon representation of TCS, with the C-terminal domain colour-coded green, is shown on the right panel. Residues with large changes in amide chemical shifts (as in Figure 3c) are colour-coded magenta. (b) The structure of eEF2–SRL complex was derived from an 11.7-Å cryo-electron microscopy map of the 80S ribosome complexed with eEF2 and sordarin (38) (PDB code 1S1H and 1S1I). Residues Q176-T191 (colour-coded dark green) of eEF2 were found to be in contact with P-proteins in the cryo-electron microscopy map (38). (c) The model of TCS–SRL complex was obtained as described in the Materials and methods section. The C-terminal domain of TCS is colour-coded magenta. (d) The model of TCS–SRL complex is superimposed onto the structure of eEF2–SRL complex. The three basic residues (K173, R174 and K177) that were found to be involved in binding P2 are shown in ball-and-stick representation in (c) and (d). Noteworthy, the P-protein-binding site (dark green) on eEF2 is in close proximity to the P-protein-binding site (magenta) of TCS.