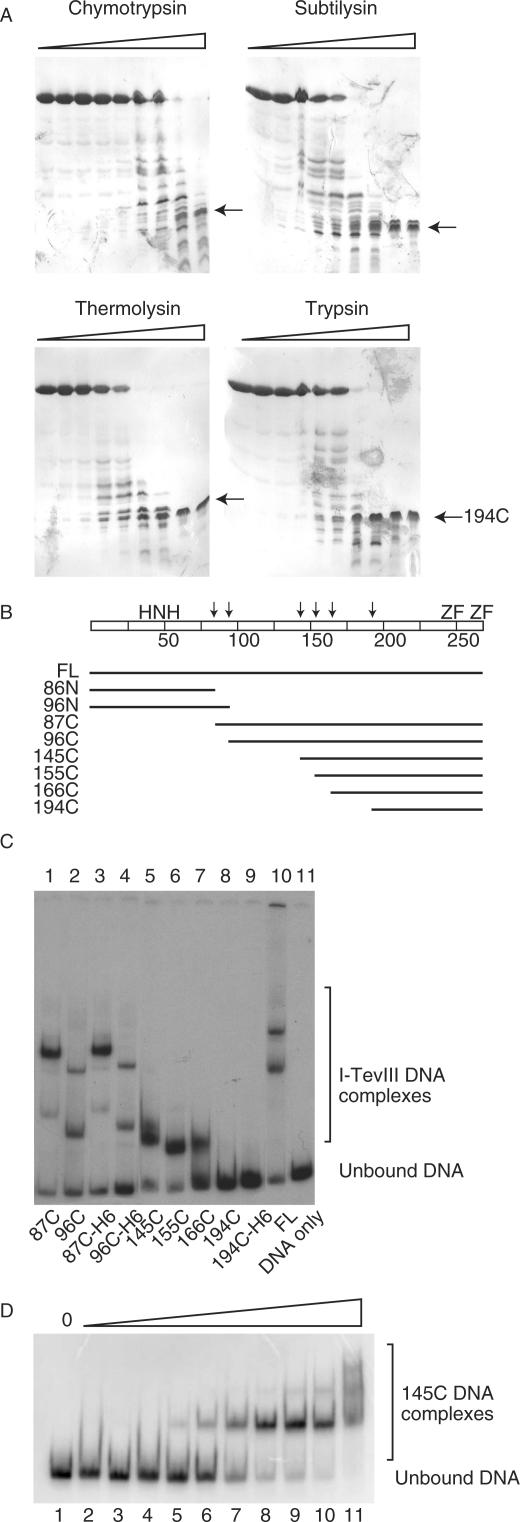
Figure 4.
I-TevIII analysis by limited proteolysis. (A) Representative SDS-PAGE gels of I-TevIII digested with proteases chymotrypsin, subtilysin, thermolysin and trypsin. The arrows indicate a proteolytically stable fragment of ∼7 kDa. (B) Schematic of protease-sensitive sites and derivatives constructed based on those sites. FL = full length. The arrows indicate proteolytically sensitive sites, and the scale of the bar is calibrated in 25-amino-acid intervals. The H–N–H motif and zinc fingers (ZF) are marked. (C) DNA-binding activity of I-TevIII and deletion derivatives. Samples were separated on native 8% polyacrylamide gels. (D) DNA-binding activity of 145C. There was a two-fold increase in the amount of protein per lane, starting with 0.001 µg and ending with 0.512 µg.