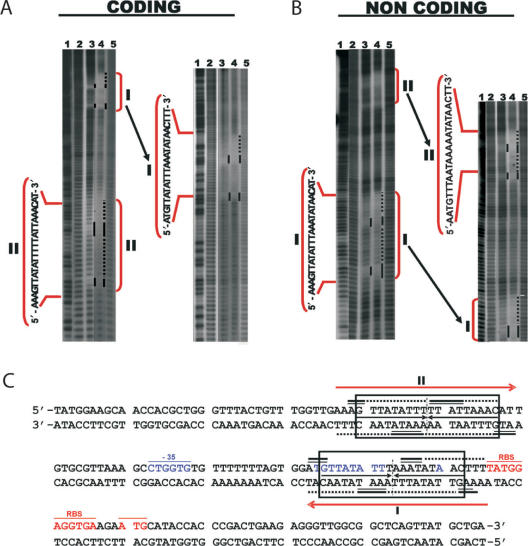
Figure 2.
Kid–Kis and Kis interact at specific sites with parD DNA. Hydroxyl radical footprinting assays were performed on Kid–Kis mixtures (Kid–Kis ratio 1:2; Kid 2.4 and Kis 4.8 µM) and Kis (4.8 µM) alone on the 175-bp parD region I/II fragment. Protections in the coding (A) and non-coding (B) strands are indicated by black bars and dots. Lane 3 shows the protection pattern by Kis alone and lane 4 shows the protection pattern by the Kid–Kis complex. The sequences of the inverted repeats I and II that include the protected regions are indicated. Lanes 2 and 5 show the cleavage pattern of the DNA in the absence of any added protein. Lane 1 shows the Maxam–Gilbert AG ladder sequence. (C) Summary of the protected sites in parD region I/II by Kis and Kid–Kis complexes. The protected regions are indicated with numbers I and II. Region I contains an 18-bp perfect two-fold symmetry element (boxed) that includes the −10 motif. The site II includes an 18-bp pseudo-symmetric element that is also boxed. The dyad symmetry axis in each region is indicated with a broken line. Bases whose deoxyriboses are protected by Kis (thick bars) or Kid–Kis (thin bars) from cleavage by hydroxyl radical are indicated (underlined). DNA sequence of the −35 (underlined and labelled) and −10 elements of the promoter and the transcription initiation site are in blue. The ribosome-binding site (RBS) and translation initiation condon (Met) of kis are underlined and in red. The imperfect inverted repeats I and II are indicated with red arrows. The 30-bp DNA fragment used for mass spectrometry studies contains parD region I plus 2 bp upstream and 5 bp downstream.