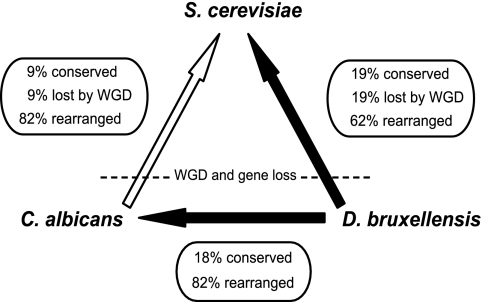
FIG. 3.
Estimated rates of chromosomal rearrangements between D. bruxellensis, C. albicans, and S. cerevisiae. Black arrows indicate analyses performed in this study, while the white arrow refers to a previous study (55). The percentage of adjacent gene pairs conserved between species was calculated from the data, and the percentage of gene pair adjacencies disrupted by gene loss following whole genome duplication (WGD) in S. cerevisiae is estimated to be approximately equal to the percentage conserved. The remaining gene pairs have been disrupted by chromosomal rearrangements.