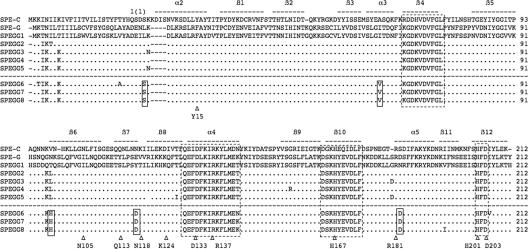
FIG. 2.
Multiple alignment of SPE-C, SPE-G, and SPEGG amino acid sequences. The amino acid sequences were aligned using Clustal W (35). The bars above the sequences indicate the secondary structure based on the crystal structure of SPE-C (PDB identification no. 1AN8). Highly conserved regions and the zinc-binding motif are boxed with dotted lines. SPEGG variants derived from isolates from animals (SPEGG6 to SPEGG8) are separated with a dotted line from those from humans, and the residues commonly mutated in SPEGG6 to SPEGG8 are indicated in boxes (also see Fig. 6). Corresponding residues of interest (see text for details) in the alignment are marked with Δ.